Benchtalk Workshop Example
Overview
The following example creates results in a demo environment.
In this example, assume we're in a lab running liquid chromatography. Scientists want to run LC, analyze the data with some third-party software, and record those data against samples that are already stored in Benchling. An example of some data is in this csv.
We will start by trying to create results in Benchling, and walk through the other APIs we will need to call along the way.
Insomnia Setup
We use a tool called Insomnia to make HTTP requests to Benchling's API. You can install Insomnia at https://insomnia.rest, and follow along by importing this configuration.
You can import configuration using the "Import/Export" option shown below.
All the imported requests will use Basic HTTP auth with your api key. To access this yourself (or set it for new requests), you can open the second tab of any request.
Benchling API Calls
Creating Results
Once Insomnia is set up, open the first request. This is an empty request to create results.
The body of this request has a list of assayResult objects, which each have two top-level keys: one for the schema of the result, and one for a dictionary of all the fields associated with it.
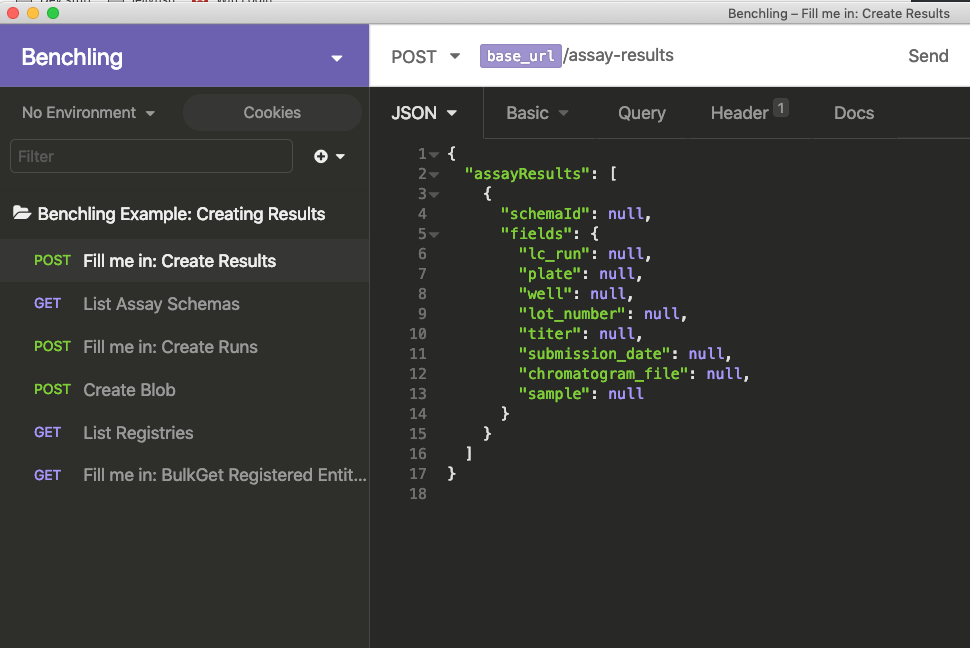
Strictly speaking, the schema ID is the only property you need. However, you probably want to fill in at least some of the fields for your results as well (and, depending on your result schema, some of these fields may be required).
Getting your result schema ID
To fill in the result schema ID, you will list all result schemas, and pick the one that you want. Result schemas do not have their own endpoint; instead, one endpoint (GET /assay-schemas) lists schemas for both results and runs. If you run the second request, you'll see all available schemas on the far right.
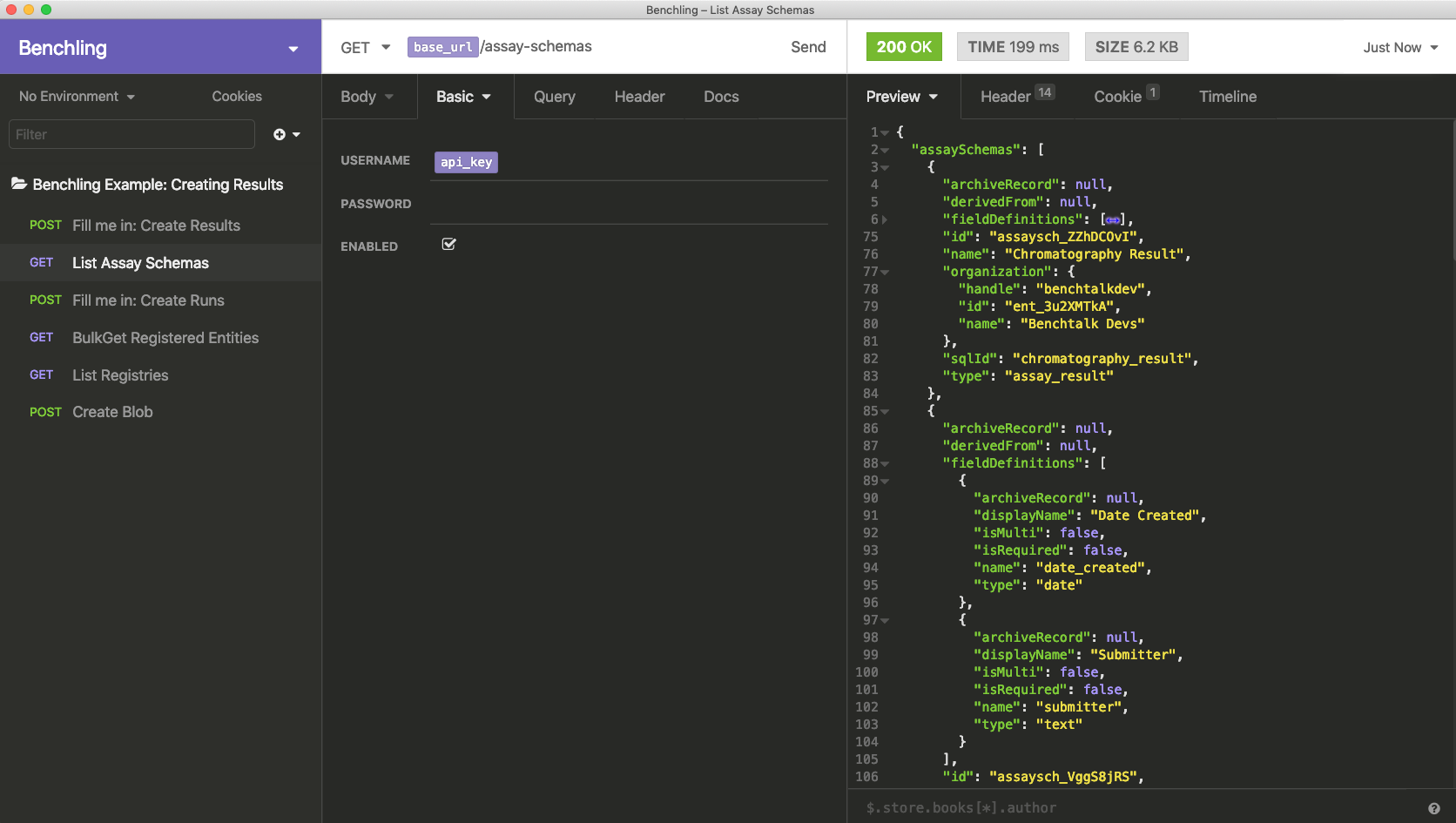
In this case, we're looking for the "Chromatography Result" schema. We'll find that object, copy its id property, and paste that into the schemaId property of the Create Results request above.
This request is usually something we'll just need to do once; after that, we can store the values we get as constants that our integration can use.
Creating a run
Runs in Benchling represent runs of an instrument. Typically a run will contain many results, measured against many different samples. While they aren't necessary to upload results, it makes managing them afterwards much easier.
To create a Run, open the Create Runs request. As with results, you will need the ID of the schema you want to use, along with any fields.
In our example, there is only one field to fill in, and it's text. The schema can be found in the GET /assay-schemas response from earlier. This time, instead of looking for "Chromatography Result", we look for "Chromatography Run". The below image shows this information filled out.
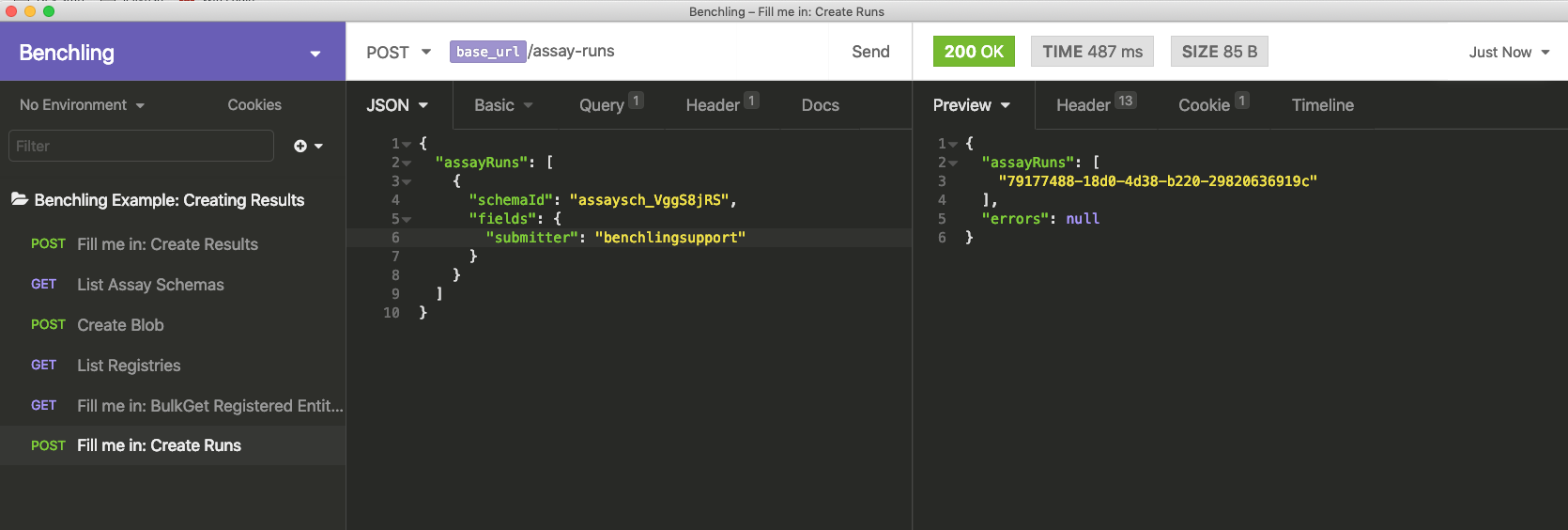
The ID of the run is returned from this request, and should be copied for the next step.
Fill in fields
After we fill in the schema ID, we can also fill in most fields.
The first field is a run link. We'll talk more about link fields later, but for now, paste in the run ID returned from the last step.
The next five fields shown all have primitive types (text, float, and date in this case). We can just fill these in with data from the spreadsheet of results, so we'll have something like the below.
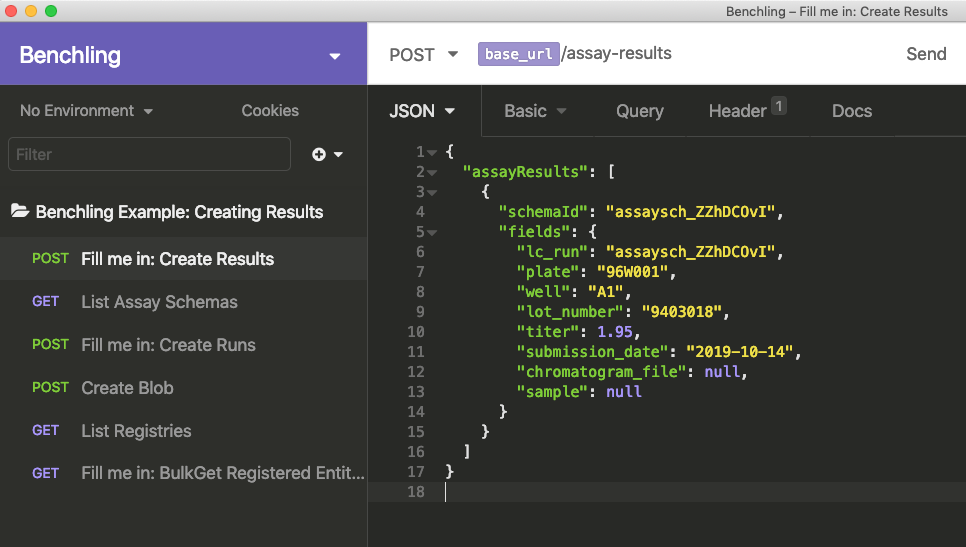
The last two fields are link types. To fill in these, we'll need to look up or create the data we want to link to, copy the ID, and use that as the value.
The field definitions of the link fields are shown below (this is what we get from the GET /assay-schemas endpoint. Notice that the type property of all three is some kind of link. The name of this indicates what kind of object we need to link to.
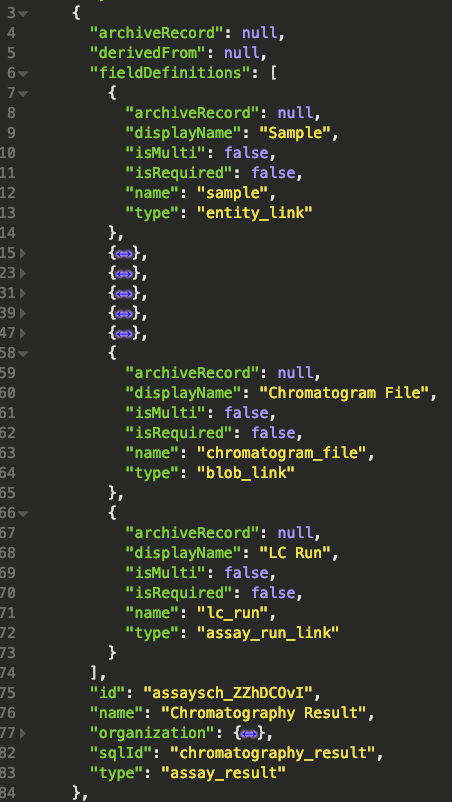
Uploading the chromatogram file
The first link field to fill in is chromatogram_file. This is a blob link, which is useful for uploading things like images.
To create our own blob link, we'll use the POST /blobs endpoint. The Insomnia template has this endpoint pre-filled with an example graph from a chromatography analysis tool.
This takes a base64-encoded file, along with some metadata, and creates a file in Benchling. A unique ID is returned, which we can paste into the appropriate part of the Create Results endpoint.
Looking up an entity to link the result to
Next, we'll want to link to the actual sample that was being measured. The ID in the spreadsheet is one that was generated for a registered entity.
First, we need to find the registry that the sample was registered into. This can be done by listing all registries with the GET /registries endpoint, and finding the registry you're interested in. Run this, and copy the ID of the registry. If this was part of a larger integration, this ID could be saved as a constant, and referenced throughout.
Second, we need to use the GET /registered-entities:bulk-get endpoint to look up the specific entity with the appropriate registry ID. This endpoint requires the registry ID to be filled in as a path parameter (the ID you just found), as well as a query parameter for entityRegistryIDs. A filled-out version is shown in the image below. In the Insomnia template, you will need to replace <registry_id> in the path with your registry ID.
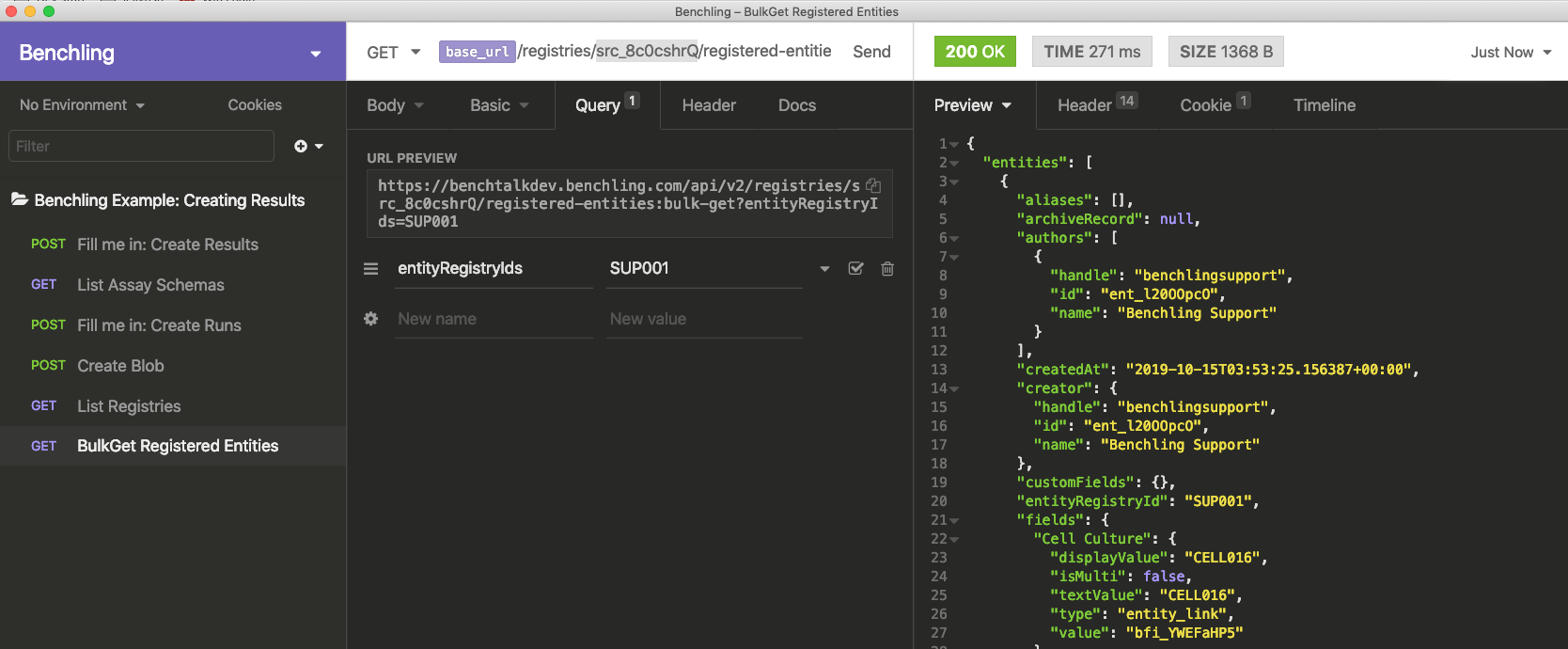
From here, copy the id of the returned entity, and paste it into the value for the sample property of the result (Note: id is not the registry ID. id is a system-generated random ID that the API uses.)
Creating results (again)
After pasting in your run ID, you'll have all the data you need to fill out your Create Results request (as shown below). Click "Send", and you'll be all set.
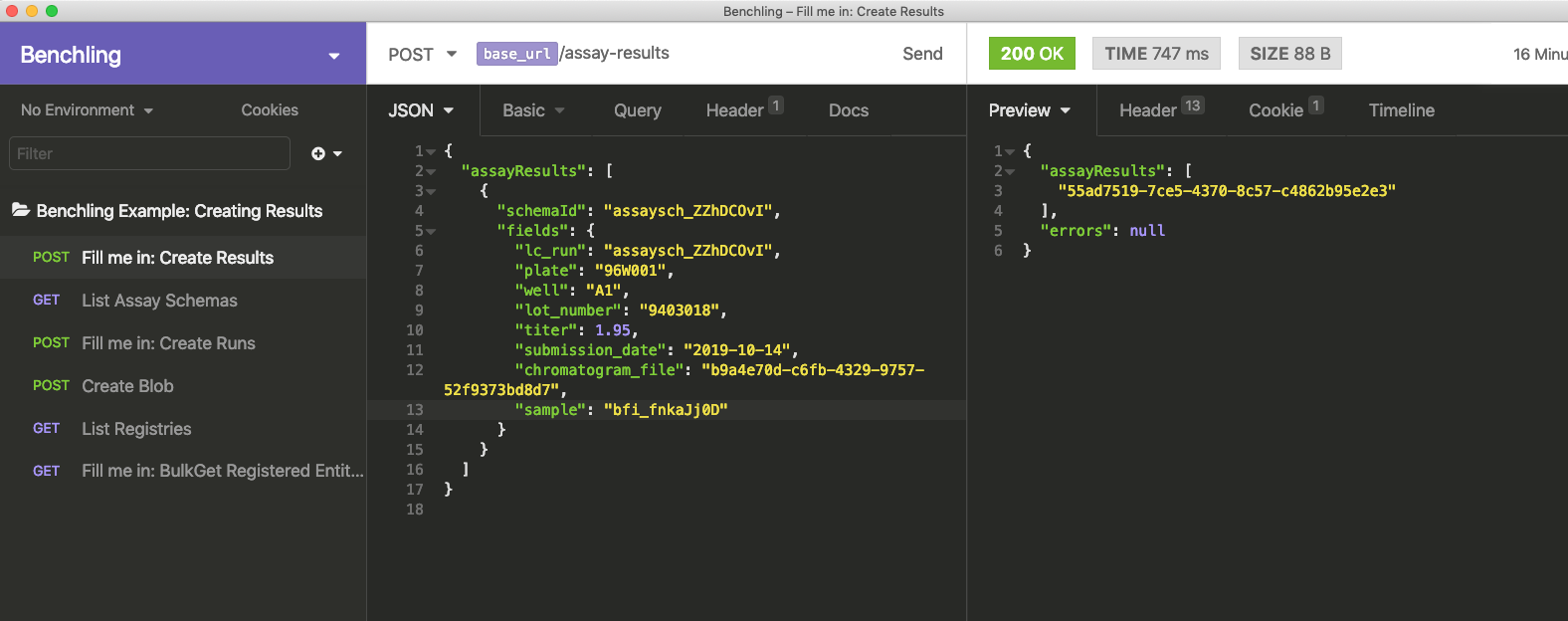
Results created this way can be inserted into the Benchling Notebook, by inserting a run, and choosing "Insert from Inbox". This will let you see API-uploaded runs. Once you select it, all results uploaded to the run will appear.
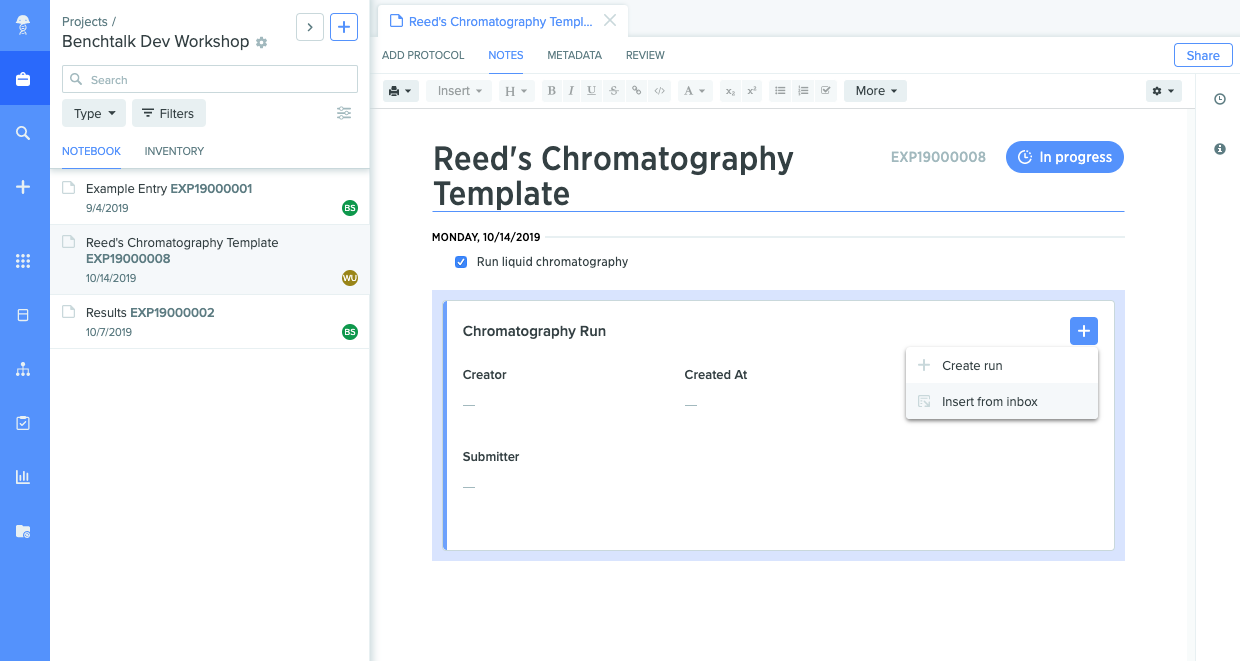
After this, you're all set! Once you're comfortable making these calls in a language of your choice, you can try writing a tool to iterate through a CSV and make them for each line in it.
Updated over 4 years ago
