Building your first app with App Configuration
Building your first app with App Configuration
For the purposes of this tutorial, we’ll use the simple app we described in Building your first App. To review, this app updates the “species” field for all DNA sequences of a certain schema. The full code is included here for reference:
from benchling_sdk.benchling import Benchling
from benchling_sdk.models import DnaSequenceBulkUpdate, AsyncTaskStatus
from benchling_sdk.auth.client_credentials_oauth2 import ClientCredentialsOAuth2
from benchling_sdk.helpers.serialization_helpers import fields
from datetime import datetime, timedelta
import random
tenant_name = "Your tenant"
client_id = "Your App's Client ID"
client_secret = "Your App's Client secret"
auth_method = ClientCredentialsOAuth2(
client_id=client_id,
client_secret=client_secret,
token_url=f"https://{tenant_name}.benchling.com/api/v2/token")
benchling = Benchling(url=f"https://{tenant_name}.benchling.com", auth_method=auth_method)
# Update this value to specify which schema this script will poll
entity_schema_to_check = "ts_AcqPy93F"
# Generates yesterday's date as a string formatted in YYYY-MM-DD
today = datetime.today().date()
yesterday = today - timedelta(days=1)
time_string = f"> {yesterday.strftime('%Y-%m-%d')}"
species_list = ["Human", "Mouse", "Rat", "Camel"]
# This creates a generator that returns DNA sequences that match the filter params provided
dna_sequences_list = benchling.dna_sequences.list(modified_at=time_string,schema_id=entity_schema_to_check,page_size=100)
def update_species_bulk(sequence_pages):
# We now loop through the pages provided by the generator and update the Species field as required
for page in sequence_pages:
updates = []
for sequence in page:
current_species = sequence.fields["Species"].value
if not current_species:
species_to_add = random.choices(species_list)[0]
update = DnaSequenceBulkUpdate(
id=sequence.id,
fields=fields(
{
"Species": {"value": species_to_add}
}
)
)
updates.append(update)
if updates:
# Calls the Bulk Update API and returns an AsyncTask we need to poll
update_task_link = benchling.dna_sequences.bulk_update(updates)
# Polls every 1 second up to 30 seconds by default to see if it completes
update_task = benchling.tasks.wait_for_task(update_task_link.task_id)
if update_task.status != AsyncTaskStatus.SUCCEEDED:
raise RuntimeError(f"Failed to update entities. Task ID: {update_task_link.task_id}\nError: {update_task.errors}")
print("Completed bulk update for task", update_task_link.task_id)
else:
print("No entities found to update")
update_species_bulk(dna_sequences_list)Consider trying to install and run this app on a new Benchling tenant; what challenges might you run into? You might have noticed some specific challenges:
entity_schema_to_check = "ts_AcqPy93F"- The app currently includes a hard-coded reference to the tenant-unique ID value for the entity schema. A user installing this app to a new Benchling tenant will have a different ID value, and might have an entirely different type of schema for which they want to autofill the “species” field.
current_species = sequence.fields["Species"].value- The app currently includes a reference to a schema field named “Species” exactly. In a different tenant, the field names are unlikely to be identical; for example, the field name might be “Organism” instead of “Species”.
We can leverage an app manifest to remove these tenant-specific dependencies from our app . First, let’s create a YAML file and build a manifest that includes configuration options to capture these details:
manifestVersion: 1
info:
name: My App Name
description: A user understandable description of what the app does, and support contact info.
configuration:
- name: DNA Sequence Schema
description: Select a DNA sequence schema with a species field for this app to autofill
type: entity_schema
fieldDefinitions:
- name: Species
description: Field that the app will write the species into.You’ll see that we defined two configuration options: One field of type entity_schema, and another text field with the name Species. Our goal is to update our app to reference these fields instead of the tenant-specific identifiers, allowing Benchling users to update the fields with the correct values. To this end, we need to install our app using this manifest:
The Benchling App Workspace
The app settings page for an example app
Now that we’ve created the app from the manifest, we can follow the steps discussed under Distributing an App in App Workspace & Configuration guide to complete the installation process. With that complete, we can finish configuring the app. Navigate to the app’s configuration page by clicking the “Connections” sidebar option → “Apps” tab → “My App Name” → “Configuration” tab:
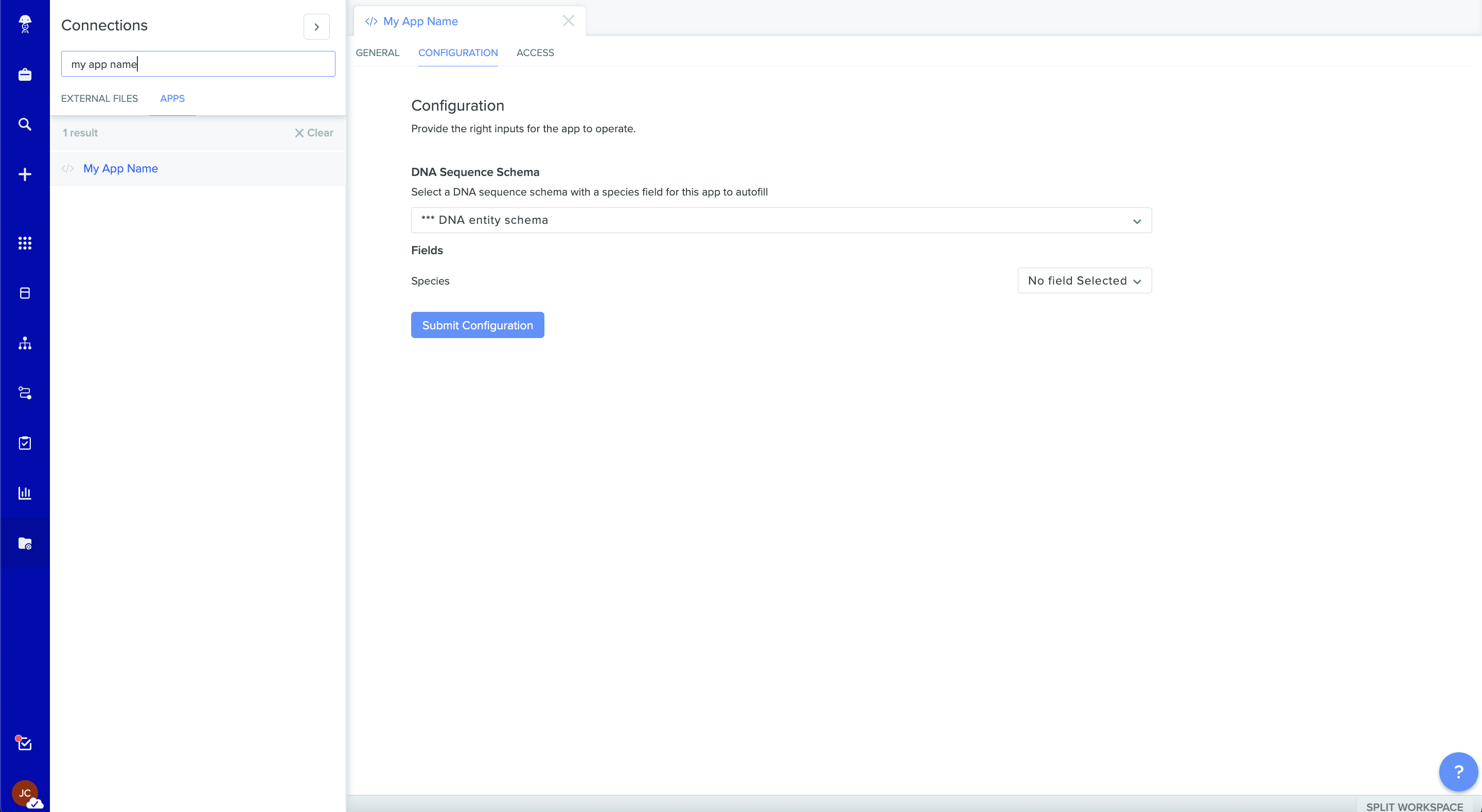
The app configuration page for our example app showing our configuration fields
Once there we can see that our schema configuration options, defined in the App Manifest, can be configured from our App Configuration page. Select the schema on this tenant that we want this app to interact with using the schema selector:
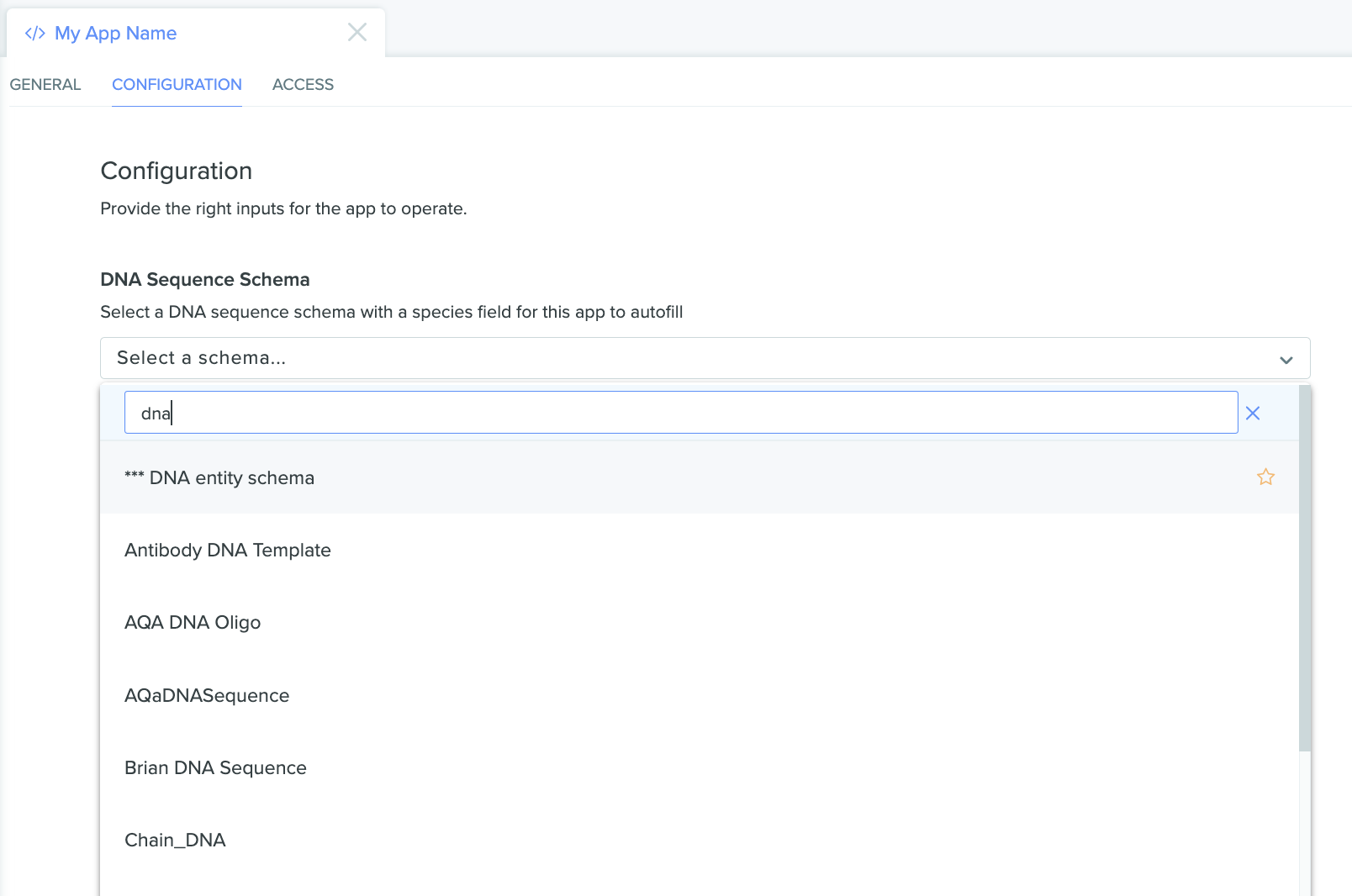
The app configuration page with an open dropdown menu listing entities that can be selected for our configuration field
Once the appropriate schema is selected, select the field on that schema we want the app to use, and then submit the configuration:
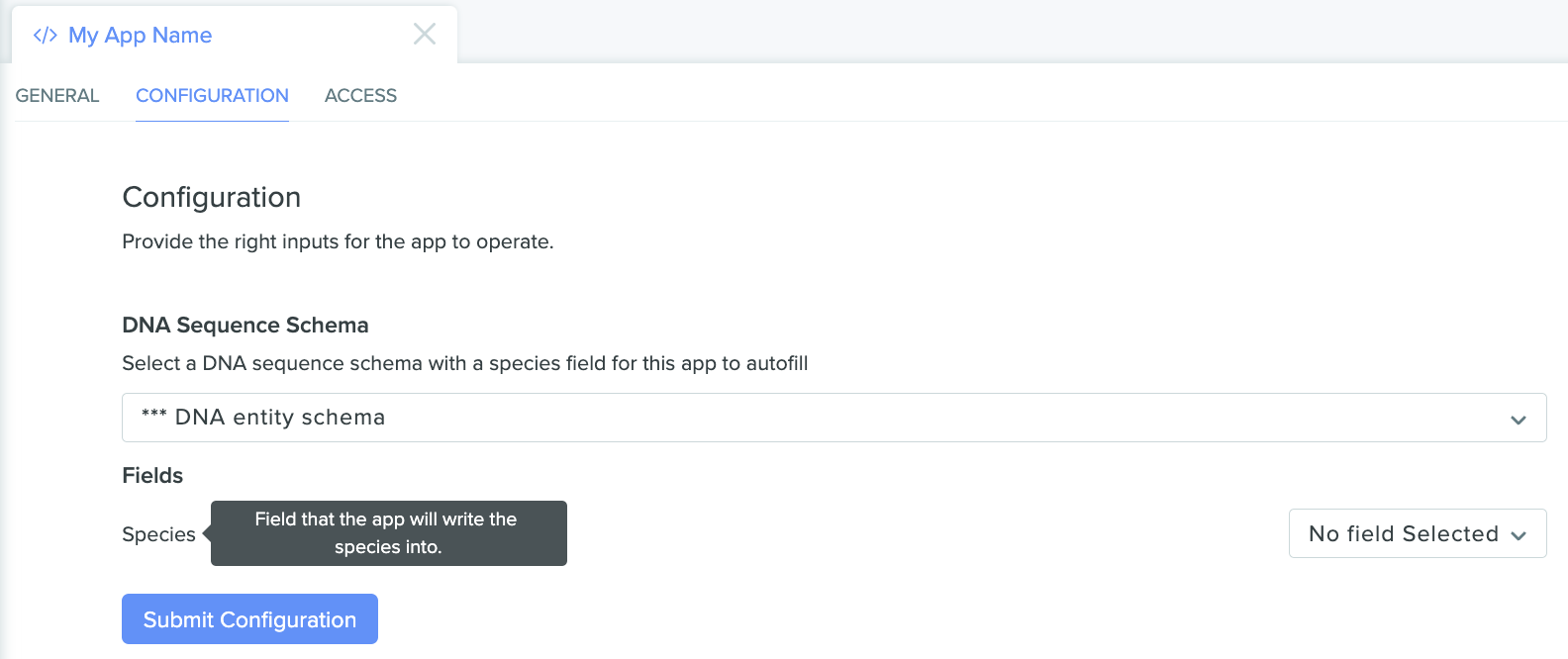
The app configuration page showing our selected entity schema with an additional "Fields" selector
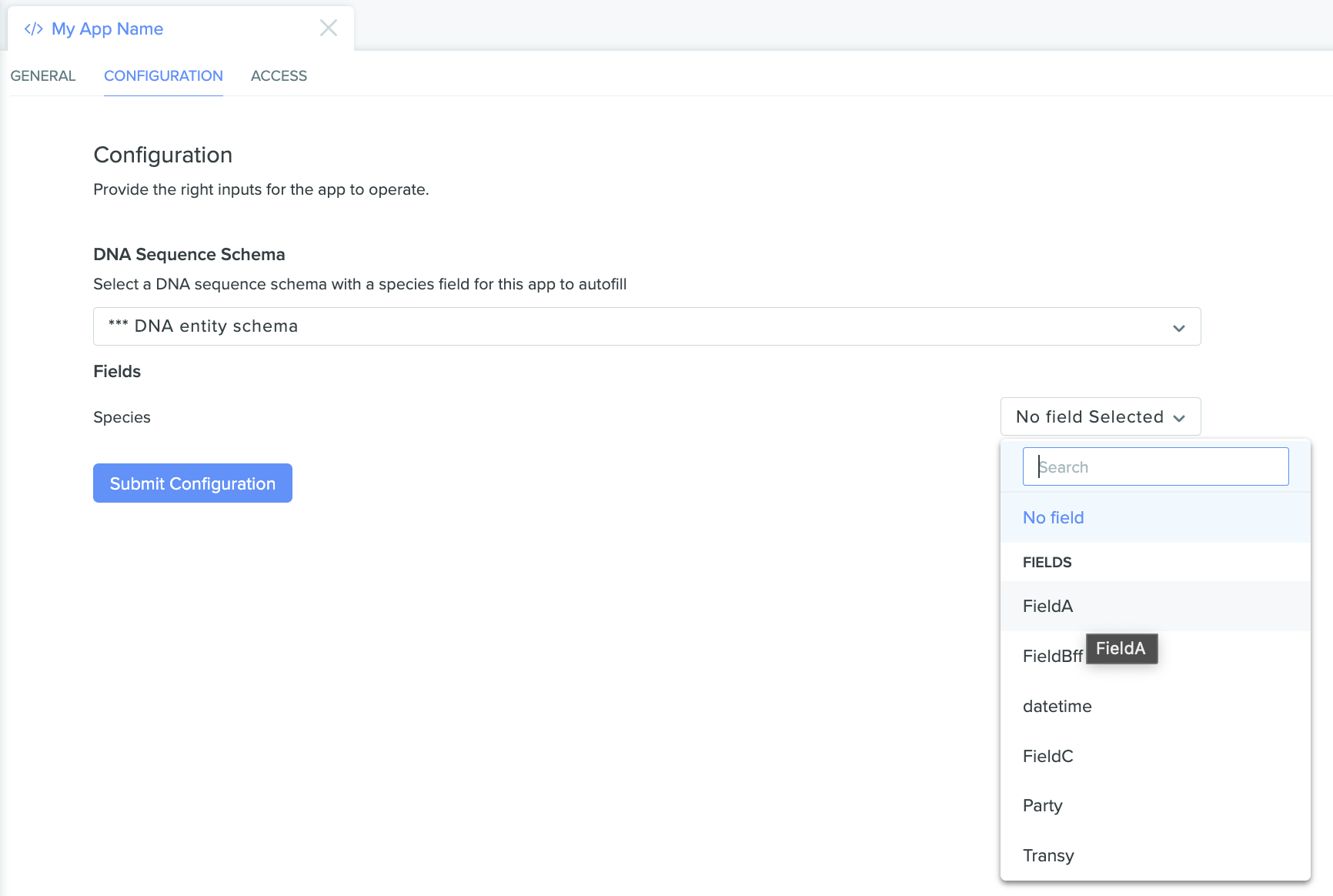
The app configuration page showing our selected entity schema with an additional "Fields" selector, this time with an open dropdown listing options
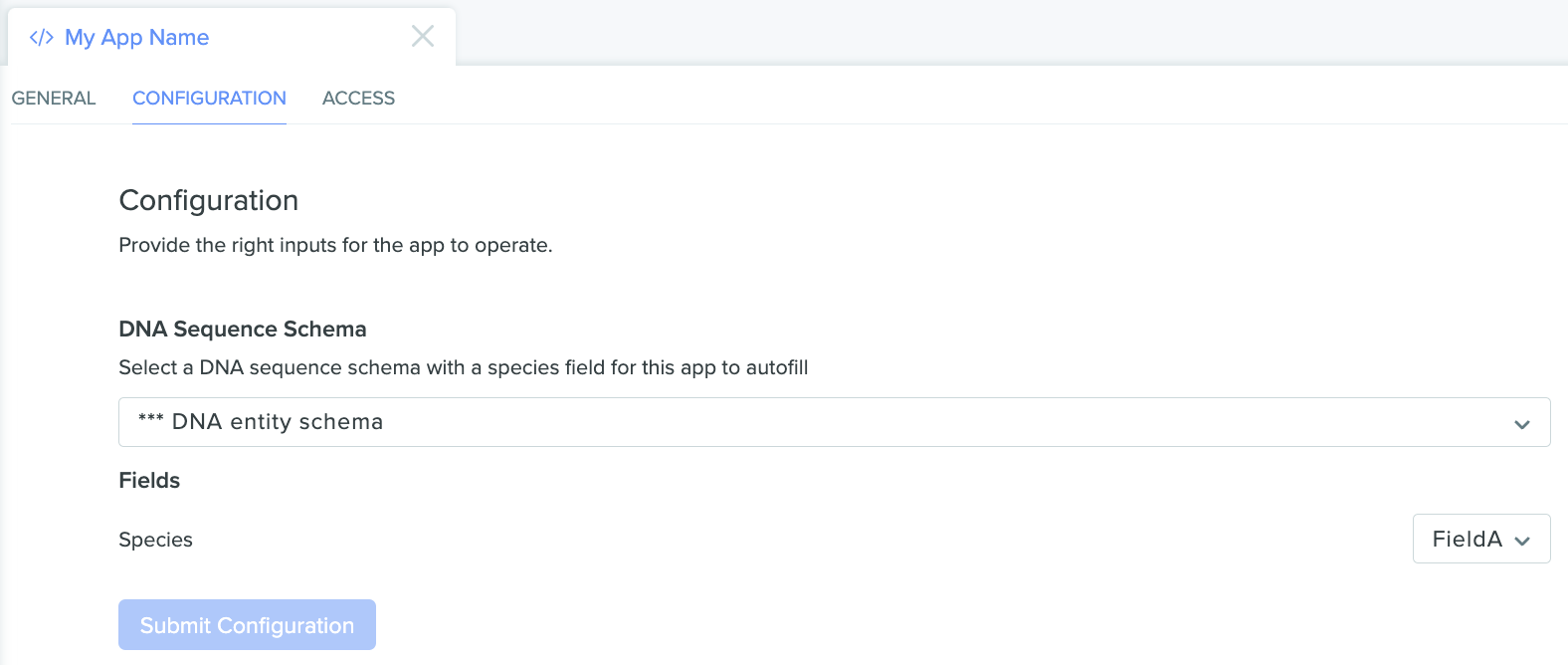
The app configuration page showing our selected entity schema and our selected field
While we are performing this process ourselves for the purposes of this tutorial, the power of App Configuration is that a tenant admin can perform these configuration steps themselves. This allows you to build tenant-agnostic apps that can be installed by multiple customers.
Next, we’ll look at how to update our code to leverage the new App Configuration.
Updating app code to use Configuration
The example app we’ve been working with has values for tenant URL, credentials, and app ID hard coded as variables for demonstration purposes. This isn’t a best practice, and storing these in a secure parameter store accessible by your app is required to use App Configuration. We’ve omitted the details of that process here, since it’s unique to your app’s deployment environment.
Using the credentials for the app, the app ID, and the tenant URL, apps can access their own configuration through the Get app configuration items endpoint. The response from this endpoint will look like this:
{
"appConfigurationItems": [
{
"apiURL": "https://piproduct.benchling.com/api/v2/app-configuration-items/aci_ksU0sJ8w",
"app": {
"id": "app_kcWboNOQCnKjFfGQ"
},
"description": "Select a DNA sequence schema with a species field for this app to autofill",
"id": "aci_ksU0sJ8w",
"linkedResource": {
"id": "ts_R3UNHyel",
"name": "*** DNA entity schema"
},
"modifiedAt": "2022-10-25T22:58:12.793897+00:00",
"path": [
"DNA Sequence Schema"
],
"requiredConfig": false,
"type": "entity_schema",
"value": "tsf_AnUZB7nO"
},
{
"apiURL": "https://piproduct.benchling.com/api/v2/app-configuration-items/aci_yM8hXMed",
"app": {
"id": "app_kcWboNOQCnKjFfGQ"
},
"description": "Field that the app will write the species into.",
"id": "aci_yM8hXMed",
"linkedResource": {
"id": "tsf_kng5zz63",
"name": "Species"
},
"modifiedAt": "2022-10-25T22:58:12.793897+00:00",
"path": [
"DNA Sequence Schema",
"Species"
],
"requiredConfig": false,
"type": "field",
"value": "tsf_AnUZB7nO"
}
],
"nextToken": ""
}The configuration options we updated earlier in Benchling are accessible to the app via the configuration endpoint. Notably, the top-level appConfigurationItems is an array containing objects that correspond to all configuration options. Each configuration item includes metadata and the configuration values selected by the tenant admin.
Specifically, we're interested in the linkedResource field, which is an object including the API ID and name values that we had previously hard coded into our app. We can update our app code to pull this configuration dynamically when we start the app, picking up relevant configuration before running:
from benchling_sdk.benchling import Benchling
from benchling_sdk.models import DnaSequenceBulkUpdate, AsyncTaskStatus
from benchling_sdk.auth.client_credentials_oauth2 import ClientCredentialsOAuth2
from benchling_sdk.helpers.serialization_helpers import fields
from datetime import datetime, timedelta
import random
tenant_name = "Your tenant"
client_id = "Your App's Client ID"
client_secret = "Your App's Client secret"
auth_method = ClientCredentialsOAuth2(
client_id=client_id,
client_secret=client_secret,
token_url=f"https://{tenant_name}.benchling.com/api/v2/token")
benchling = Benchling(url=f"https://{tenant_name}.benchling.com", auth_method=auth_method)
### Pull app config
# First define the variables we care about
entity_schema_to_check = ""
species_schema_field_name = ""
# Note: list_app_configuration_items() returns a pageIterator,so we'll
# cast it as a list to get all pages up front
config_pages = list(benchling.apps.list_app_configuration_items(app_id="app_kcWboNOQCnKjFfGQ"))
# Iterate through all pages and config items
for page in config_pages:
for item in page:
# Marshal the path property into a tuple for comparison
path = tuple(item.path)
# Store relevant IDs in our previously defined variables
if path == ("DNA Sequence Schema",):
entity_schema_to_check = item.linked_resource.id
if path == ("DNA Sequence Schema","Species"):
species_schema_field_name = item.linked_resource.name
# Generates yesterday's date as a string formatted in YYYY-MM-DD
today = datetime.today().date()
yesterday = today - timedelta(days=1)
time_string = f"> {yesterday.strftime('%Y-%m-%d')}"
species_list = ["Human", "Mouse", "Rat", "Camel"]
# This creates a generator that returns DNA sequences that match the filter params provided
dna_sequences_list = benchling.dna_sequences.list(modified_at=time_string,schema_id=entity_schema_to_check,page_size=100)
def update_species_bulk(sequence_pages):
# We now loop through the pages provided by the generator and update the Species field as required
for page in sequence_pages:
updates = []
for sequence in page:
current_species = sequence.fields[species_schema_field_name].value
if not current_species:
species_to_add = random.choices(species_list)[0]
update = DnaSequenceBulkUpdate(
id=sequence.id,
fields=fields(
{
species_schema_field_name: {"value": species_to_add}
}
)
)
updates.append(update)
if updates:
# Calls the Bulk Update API and returns an AsyncTask we need to poll
update_task_link = benchling.dna_sequences.bulk_update(updates)
# Polls every 1 second up to 30 seconds by default to see if it completes
update_task = benchling.tasks.wait_for_task(update_task_link.task_id)
if update_task.status != AsyncTaskStatus.SUCCEEDED:
raise RuntimeError(f"Failed to update entities. Task ID: {update_task_link.task_id}\nError: {update_task.errors}")
print("Completed bulk update for task", update_task_link.task_id)
else:
print("No entities found to update")
update_species_bulk(dna_sequences_list)Note how the app has been updated to resolve the issues we discussed before:
dna_sequences_list = benchling.dna_sequences.list({...}schema_id=entity_schema_to_check)- Instead of a hard coded ID value, the app uses the resource ID from the App Configuration
current_species = sequence.fields[species_schema_field_name].value- Instead of the hard coded field name
"Species", the app depends on the field listed in the App Configuration
- Instead of the hard coded field name
These changes allow the app to be installed and configured on any Benchling tenant without requiring code changes. Additionally, users can make changes to field names and schemas without affecting the app’s functionality, so long as they update the corresponding values in the App Configuration.
Updated about 1 year ago
