Building your first Benchling App
This guide will walk a user through the creation and configuration of a simple Benchling App in order to highlight the standard permissions and authentication strategies for Benchling Apps. Much of the beginning of this guide will overlap with 'Getting Started with Benchling Apps', so if you've already completed those steps, skip ahead to Building the App. Once we're done, we will have an App that reviews newly created or modified DNA entities for a filled-in Species field, and updates those with missing values.
Requirements:
- Benchling Registry
- Python development environment with Benchling SDK installed locally
- Benchling Developer Platform access
- Tenant Admin permissions
Getting Access
To get started with creating your first Benchling app, first make sure you have access to the “Developer Platform” capability. Check out how to get access here: Developer Platform Access, or check out our guide on Capability Management generally: Capability Management.
Where to find apps
Apps can be found in the Feature Settings page. There are a variety of ways to get to this page, but the easiest is to navigate through your user menu as pictured on the right.
That takes you to the “Developer Console” section of the feature settings page, and opens the “Apps” settings:
Creating an app
You can then click the “Create” button in the top right to create your first app! You can always change the name and description after you’ve created it.
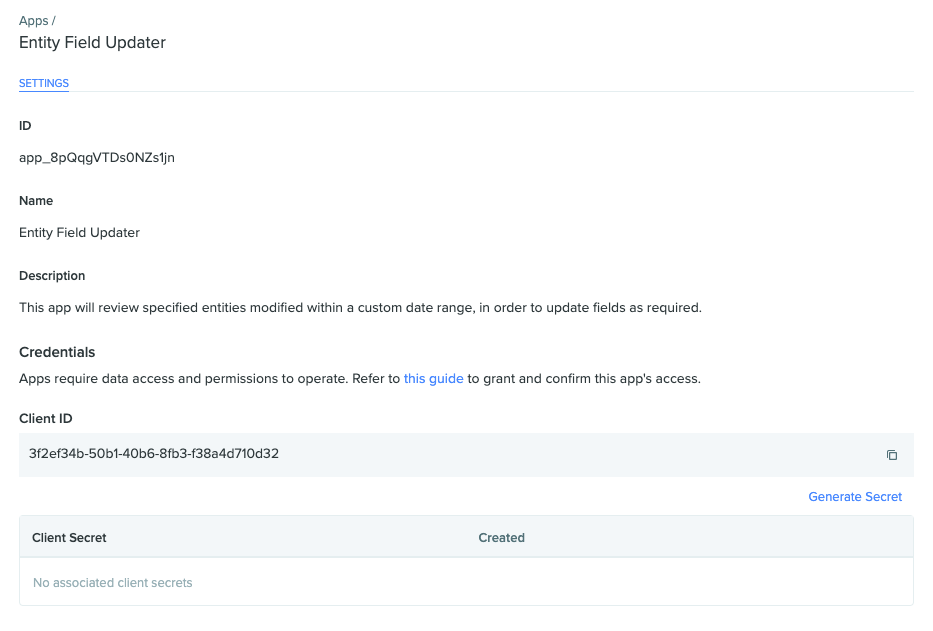
Take note of the Client ID, which is specific to the App, and save this info for future use. Next, click on the 'Generate Secret' button to create the private key for this App.
Do not close this popup yet!Make sure that you have copied the Client Secret for your App to a secure location before closing this modal window. Once you close it, the full key will not be accessible again. If you lose this private key, you will need to generate a new key, and the old key will no longer be valid.
Adding an app to an organization
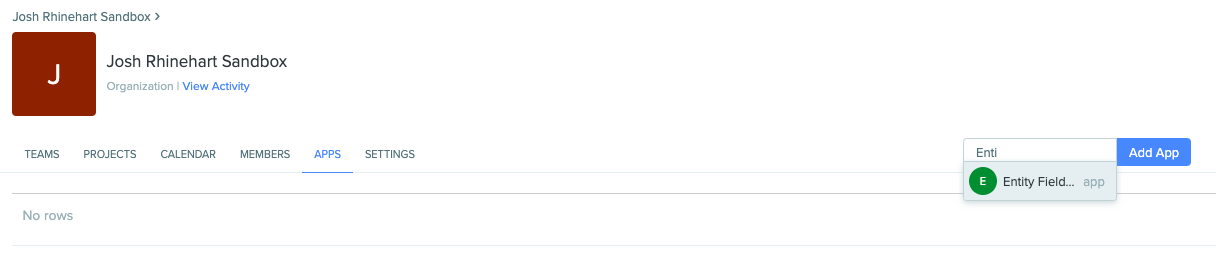
Adding an app to an organization will allow the app to access any projects that are accessible to all members of the organization as well as the Registry itself. This is generally necessary when creating a new app. Org admins can add apps in the organization page in the “APPS” tab:
Once added, it will appear in the “APPS” tab for that organization:
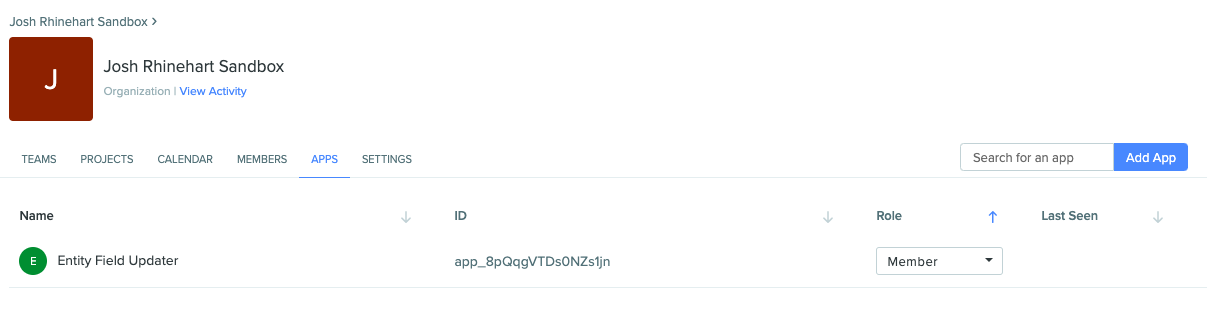
Be aware that your tenant may have been configured for Registry control by individual Projects, rather than at the Organization level. If you believe this to be the case, contact your Benchling admin and/or your Benchling Customer Success Manager for assistance.
Adding an app to Projects and Teams
Once an app is added to the organization, team or org admins can also add apps to teams, which will similarly give that app access to the projects team members have access to. This can be useful when managing several apps together (“Automation Apps”), or for apps that are specific to a certain team (“Developer Team”). For more information on these additional permissions, see the Getting Started with Benchling Apps guide.
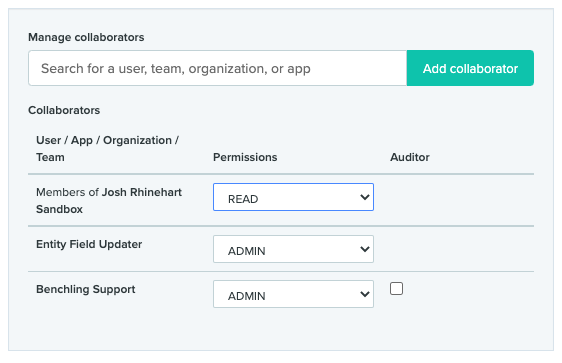
In this case, the Project housing the entities to be updated is configured so that all members of my Sandbox Org have Read-only permissions by default, so I have explicitly added the 'Entity Field Updater' App to have admin permissions within this project.
Building the App
Now that we have our App created within the Benchling platform, and the correct permissions have been granted, it's time to start building our app. Since the Benchling Registry is a highly configurable database, some App actions will necessarily be scoped to data models specific to your instance. The general purpose of this App will be to fill in missing Species data on recently created or modified DNA sequence entities, but similar logic can be used to sync with an external DB or ontology system, perform field validation, or periodically update entities with other relevant data.
We will also be leveraging the Benchling SDK, which provides a typed interface for the Benchling REST APIs. Please make sure the SDK is installed in your local environment if you would like to run this code as shown. More information regarding the SDK can be found here: https://docs.benchling.com/docs/getting-started-with-the-sdk
Data model Prerequisites
For the sake of keeping this focused on App development and not data model design, we will only require a single DNA entity schema, with a text schema field for Species, as shown below. Once you have created this entity schema, or added an appropriate field to a pre-existing entity, take note of the schema API ID, either via the URL, or the Copy API ID button if you've enabled that option in your personal settings. We will use that schema API ID to filter our API calls in the App.
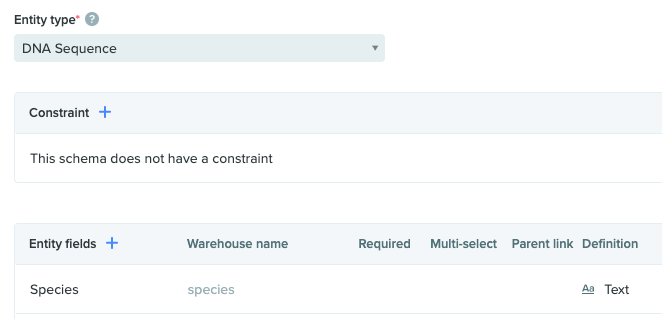
The Registry Schema configuration for a DNA sequence with a text field for 'Species'.
Authentication
If you have used the SDK in the past, then the authentication process will look very similar to your existing code. The instantiation of the Benchling client still only requires your tenant URL and auth_method, but we will use the OAuth2 Client ID and Secret key that we obtained during the App creation process rather than an individual user's API key.
from benchling_sdk.benchling import Benchling
from benchling_sdk.auth.client_credentials_oauth2 import ClientCredentialsOAuth2
tenant_name = "your_tenant"
# Your app's client credentials
client_id = "XXXXXXXX-XXXX-XXXX-XXXX-XXXXXXXXXXXX"
client_secret = "cs_AppClientSecret"
auth_method = ClientCredentialsOAuth2(
client_id=client_id,
client_secret=client_secret)
benchling = Benchling(url=f"https://{tenant_name}.benchling.com", auth_method=auth_method)Retrieving a list of DNA sequences
Now that we have our API client instantiated, we need to request a list of recently modified DNA sequences to verify. Since this list could be quite long, the SDK provides .list() methods which actually returns an iterable generator rather than the full list of entities. We can then iterate through the pages returned by this generator and take action in chunks rather than all at once. See the iterating through pages guide for more information on how the iterator works.
The list endpoint also allows for some filter parameters, so in this case we will be reducing the scope of our query to entities of the specific schema type that we are interested in, and only those with a modified_at timestamp within the last day. We've also bumped up the page size from the default of 50 to 100 to keep API calls to a reasonable level.
from datetime import datetime, timedelta
# Update this value to specify which schema this script will poll
entity_schema_to_check = "ts_AcqPy93F"
# Generates yesterday's date as a string formatted in YYYY-MM-DD
# If this were a Production App, we would also need to incorporate time zone
today = datetime.today().date()
yesterday = today - timedelta(days=1)
time_string = f"> {yesterday.strftime('%Y-%m-%d')}"
# This creates a generator that returns DNA sequences that match the filter params provided
dna_sequences_list = benchling.dna_sequences.list(modified_at=time_string,schema_id=entity_schema_to_check,page_size=100)Iterating through the returned list of Entities
Now we can begin iterating through the returned list of entities, one page at a time, and identifying which entities require an update to the Species field. If we find an entity that requires an update, we will add it to an array of entities to update, and provide the key and value of the field to update.
Thanks to the functionality of the update endpoints, we do not need to provide the details of fields that are not being updated, so we do not need to completely unpack the entity, we can simply provide the Registry ID and the fields to be changed. For the sake of simplicity, we will also just be choosing a species at random from a hardcoded array, but one could easily extend this logic to call another function or third party API at this point.
from benchling_sdk.models import DnaSequenceBulkUpdate, AsyncTaskStatus
from benchling_sdk.helpers.serialization_helpers import fields
import random
species_list = ["Human", "Mouse", "Rat", "Camel"]
def update_species_bulk(sequence_pages):
# We now loop through the pages provided by the generator and update the Species field as required
for page in sequence_pages:
updates = []
for sequence in page:
current_species = sequence.fields["Species"].value
if not current_species:
species_to_add = random.choices(species_list)[0]
update = DnaSequenceBulkUpdate(
id=sequence.id,
fields=fields(
{
"Species": {"value": species_to_add}
}
)
)
updates.append(update)Bulk Update of DNA entities and Async Tasks
Provided that we found entities with empty Species fields, we should now have an array called updates that contains objects of type DnaSequenceBulkUpdate with the id of the entity to update, and a parameter called fields which uses the serialization helpers in the SDK to properly format the new species string.
The next step (if our array of updates is not empty) is to call the DNA sequences bulk update endpoint and pass along our list of updates. All bulk operation endpoints in the Benchling API will provide a task ID rather than waiting to send a response, as some larger processing jobs can take longer than a standard timeout. When using the SDK to call these endpoints, the .bulk_update() method returns an AsyncTaskLink that we can use to easily poll the tasks API endpoint for status updates via the .wait_for_task() method. See the async tasks section of the SDK examples guide for more information.
(Note: the indentation of this block may not translate well to copy and paste, so the full reference code is provided at the bottom of this guide)
if updates:
# Calls the Bulk Update API and returns an AsyncTask we need to poll
update_task_link = benchling.dna_sequences.bulk_update(updates)
# Polls every 1 second up to 30 seconds by default to see if it completes
update_task = benchling.tasks.wait_for_task(update_task_link.task_id)
if update_task.status != AsyncTaskStatus.SUCCEEDED:
raise RuntimeError(f"Failed to update entities. Task ID: {update_task_link.task_id}\nError: {update_task.errors}")
print("Completed bulk update for task", update_task_link.task_id)
else:
print("No entities found to update")All that's left at this point is to call the function via update_species_bulk(dna_sequences_list) and monitor the output. In a production setting, we could then schedule this to run via a system automation tool like cron or a serverless environment like AWS Lambda to run on a scheduled basis, or even upgrade the app to be evented and trigger off the entity.registered event (see the events getting started guide).
Example App Full Code
As noted above, this App leverages the Benchling SDK, which provides a typed interface for the Benchling REST APIs. Please make sure the SDK is installed in your local environment if you would like to run this code as shown. More information regarding the SDK can be found here.
from benchling_sdk.benchling import Benchling
from benchling_sdk.models import DnaSequenceBulkUpdate, AsyncTaskStatus
from benchling_sdk.auth.client_credentials_oauth2 import ClientCredentialsOAuth2
from benchling_sdk.helpers.serialization_helpers import fields
from datetime import datetime, timedelta
import random
tenant_name = "Your tenant"
client_id = "Your App's Client ID"
client_secret = "Your App's Client secret"
auth_method = ClientCredentialsOAuth2(
client_id=client_id,
client_secret=client_secret)
benchling = Benchling(url=f"https://{tenant_name}.benchling.com", auth_method=auth_method)
# Update this value to specify which schema this script will poll
entity_schema_to_check = "ts_AcqPy93F"
# Generates yesterday's date as a string formatted in YYYY-MM-DD
today = datetime.today().date()
yesterday = today - timedelta(days=1)
time_string = f"> {yesterday.strftime('%Y-%m-%d')}"
species_list = ["Human", "Mouse", "Rat", "Camel"]
# This creates a generator that returns DNA sequences that match the filter params provided
dna_sequences_list = benchling.dna_sequences.list(modified_at=time_string,schema_id=entity_schema_to_check,page_size=100)
def update_species_bulk(sequence_pages):
# We now loop through the pages provided by the generator and update the Species field as required
for page in sequence_pages:
updates = []
for sequence in page:
current_species = sequence.fields["Species"].value
if not current_species:
species_to_add = random.choices(species_list)[0]
update = DnaSequenceBulkUpdate(
id=sequence.id,
fields=fields(
{
"Species": {"value": species_to_add}
}
)
)
updates.append(update)
if updates:
# Calls the Bulk Update API and returns an AsyncTask we need to poll
update_task_link = benchling.dna_sequences.bulk_update(updates)
# Polls every 1 second up to 30 seconds by default to see if it completes
update_task = benchling.tasks.wait_for_task(update_task_link.task_id)
if update_task.status != AsyncTaskStatus.SUCCEEDED:
raise RuntimeError(f"Failed to update entities. Task ID: {update_task_link.task_id}\nError: {update_task.errors}")
print("Completed bulk update for task", update_task_link.task_id)
else:
print("No entities found to update")
update_species_bulk(dna_sequences_list)Updated about 1 year ago
