Uploading a Blob to Benchling
Overview
Blobs are arbitrary files that can be uploaded to Benchling and added to schema fields and some other objects in Benchling
After a blob has been uploaded, the blob can be linked to another object such as a Result or Run to be used. Blobs are not accessible on their own, and can only be accessed from a parent object they are linked to.
Entry Attachments vs BlobsAttachments in notebook entries are separate from blobs. We currently do not have a way to upload attachments to a notebook entry via the API. We plan to support this in the future, and if you need to do so, please reach out to us at [email protected]!
Uploading a single part blob
Uploading a blob under 10MB can be done through the POST /blobs endpoint.
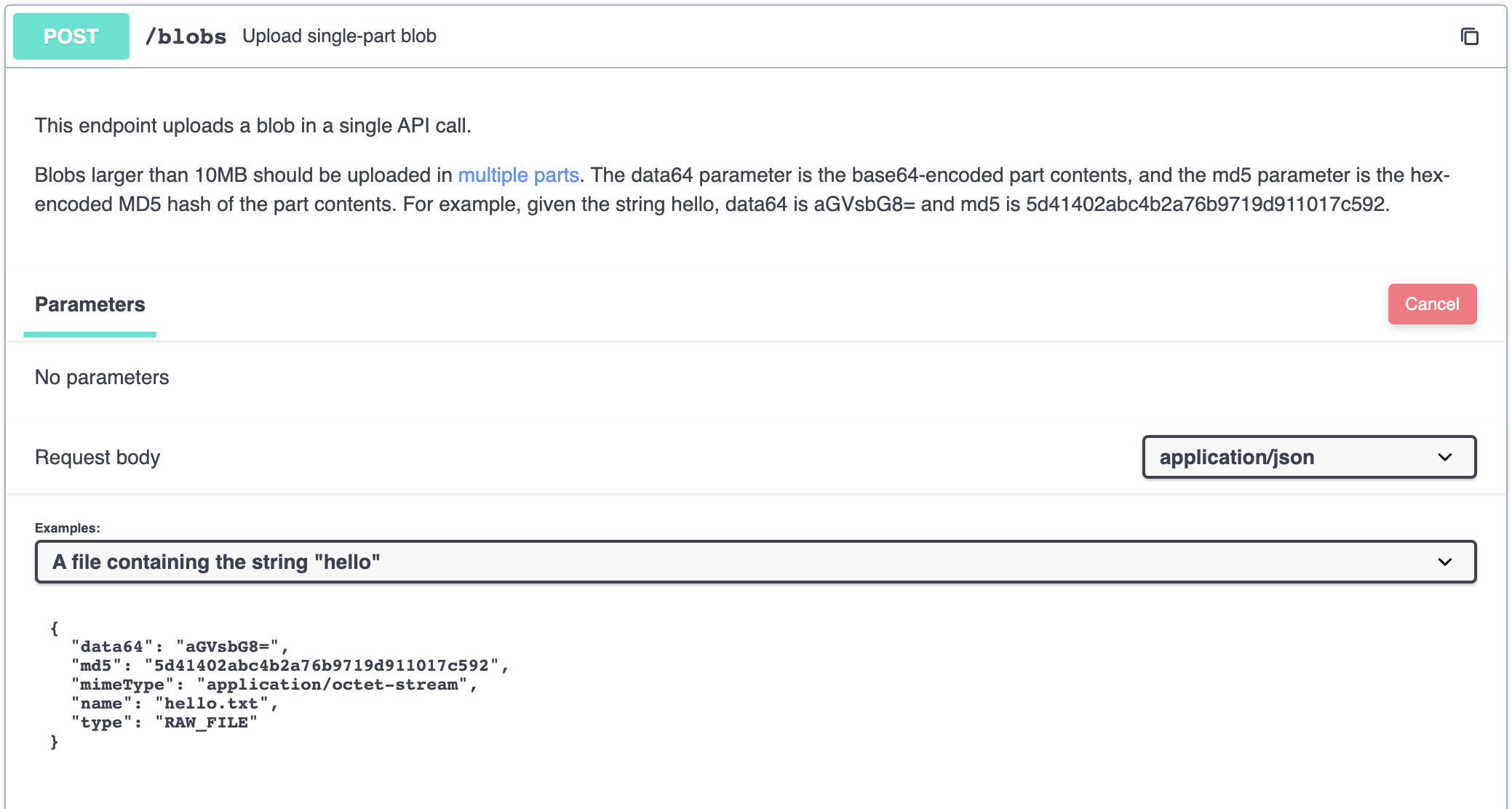
This takes a base64-encoded blob along with an MD5 hash of the original contents as part of the payload. The default mimeType is application/octet-stream.
A type of VISUALIZATION allows a preview of the image to be shown in Benchling. Other file types should denoted as RAW_FILE.
Including the base64-encoded data as part of the request can be easier to work with programmatically. Uploading a blob using python is also shown in integration-examples, where usage of uploading a single-part blob shown within upload_single_part_blob.
Store your Blob IDBlobs are not searchable on their own, so if you do not persist the ID prior to linking the blob to a parent object, it will be "lost" and you'll have to re-upload the file. Make sure to store the returned blob ID after finishing the upload so you can link it to a parent object.
Uploading a multi-part blob
If the blob is larger than 10MB, then it must be split into smaller parts, where each part (besides any final remaining part) should be at least 5MB. This workflow is shown here.
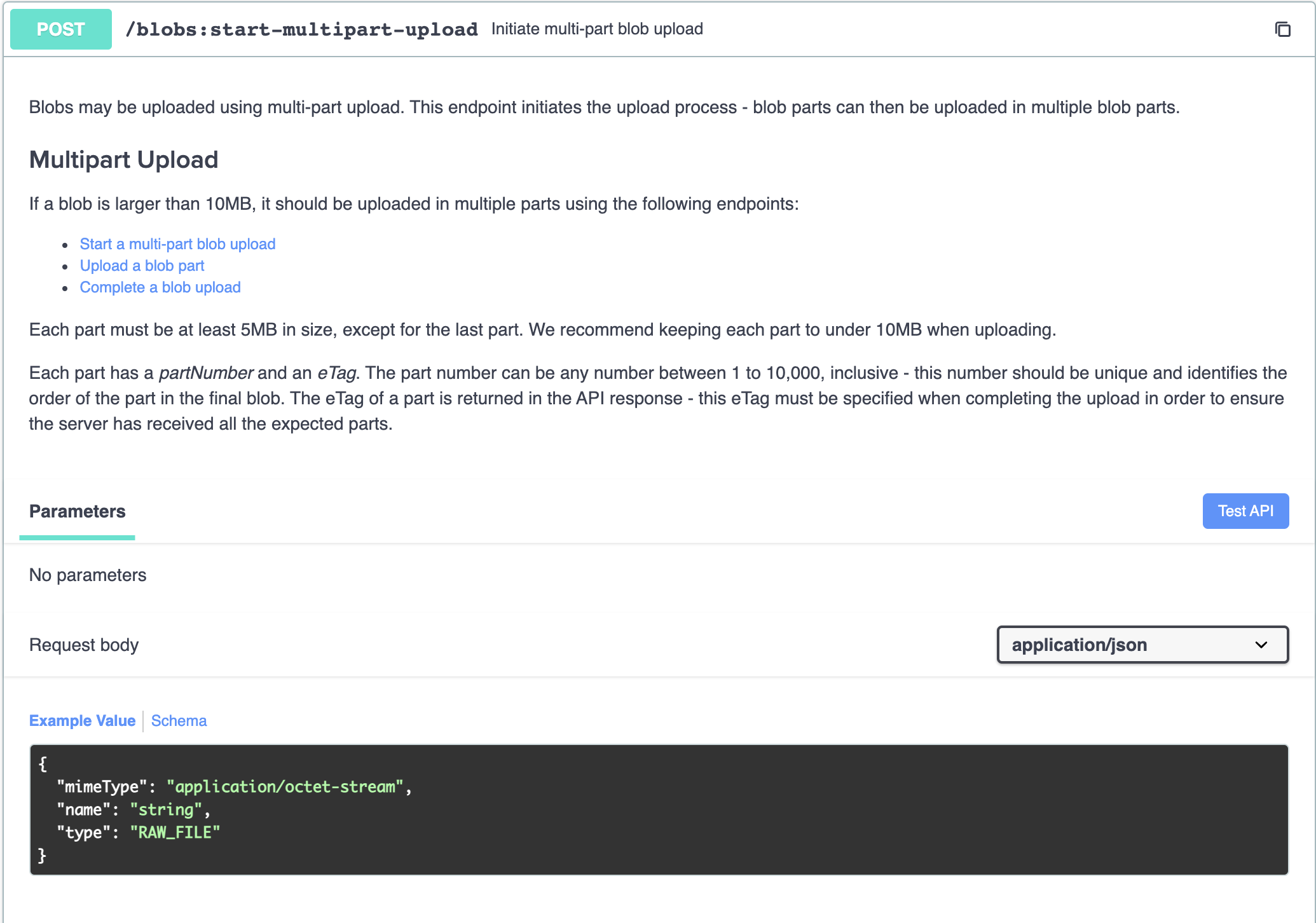
First, we initiate the multipart upload and obtain our blob ID by using POST /blobs:start-multipart-upload
After splitting our large file into 10MB parts, each part should be uploaded with an incrementing part number, beginning at 1, using POST /blobs/{id}/parts.
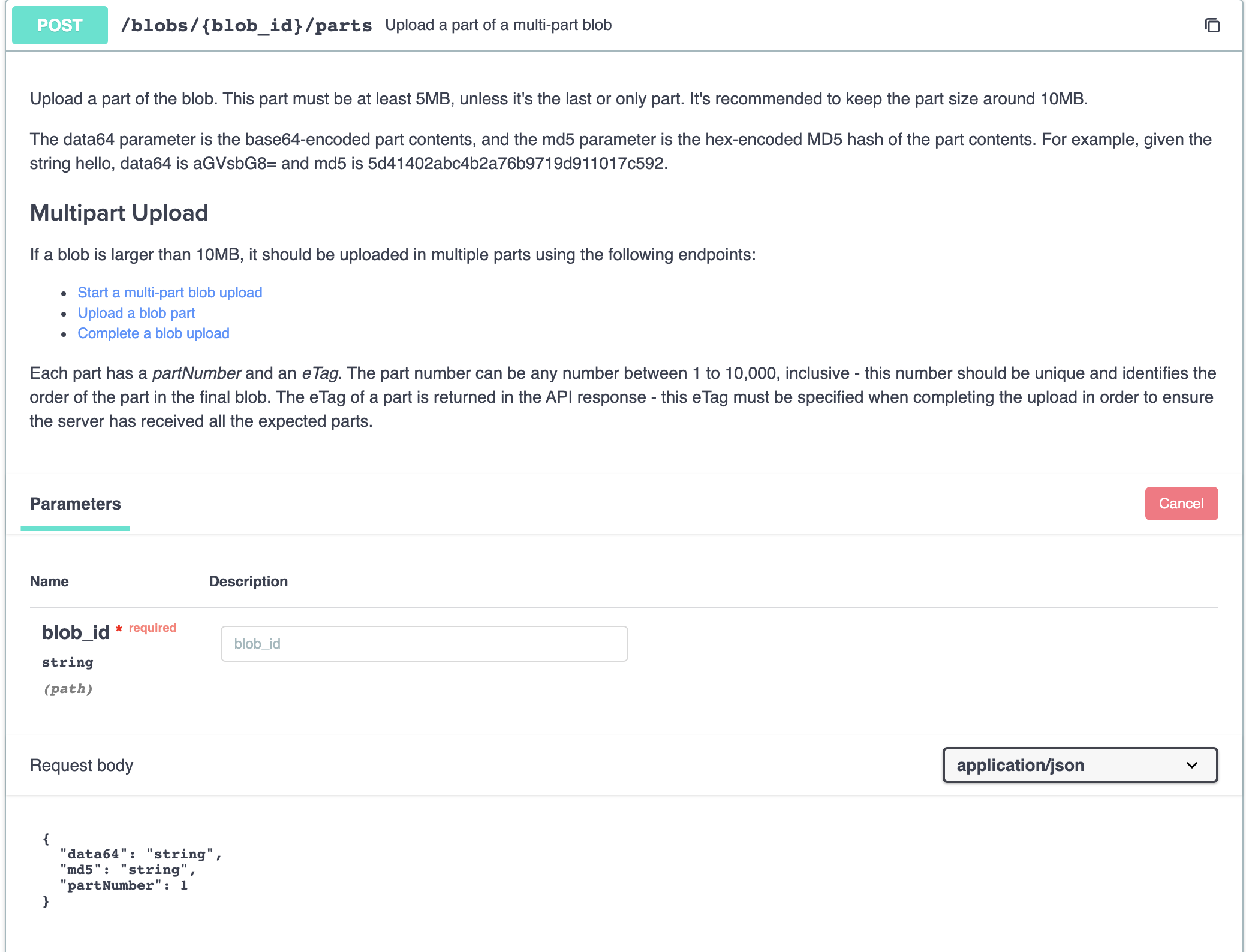
We should refer back to the response of our previous call to fill out the ID as seen in our example.
The response from this call is important to save, as to ensure the server has successfully received all parts we must send back the returned eTag and partNumber. These fields are part of the createdPart appended here.
Finally, we complete the multipart upload with POST /blobs/{id}:complete-upload.
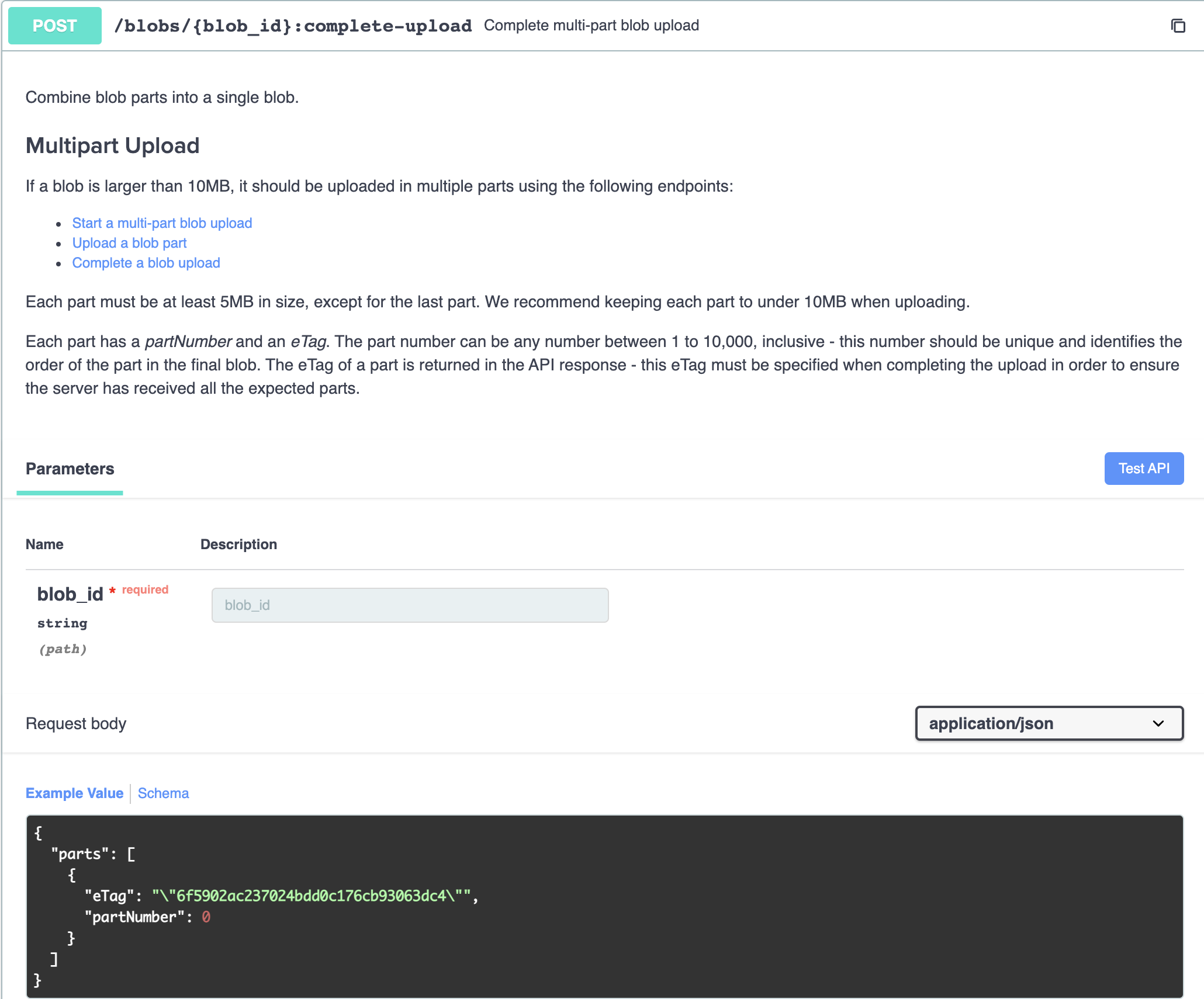
All eTags and partNumbers must be sent as part of the payload.
If any part of the upload fails, we can abort the upload with POST /blobs/{id}:abort-upload
We wrap our upload logic with exception handling that will abort the upload.
Linking Blobs to Schema Fields
Most types of schemas within Benchling support a schema field type called Attachment, programatically also referred to as blob link. These fields are where blobs are linked to objects and become accessible in Benchling.
Once a blob has been uploaded, calling the PATCH endpoint of the associated object will allow you to set the attachment field to be the blob ID that was provided upon successful upload.
Upload the Blob
For this example, we'll upload our example blob so we can link it to a schema field. First, construct the request body for the single part blob upload:
{
"data64": "iVBORw0KGgoAAAANSUhEUgAAAB4AAAAUBAMAAABohZD3AAAAG1BMVEUAAAD////f39+/v78fHx9/f39fX1+fn58/Pz+3dh1rAAAACXBIWXMAAA7EAAAOxAGVKw4bAAAAT0lEQVQYlWNgoDlQBhGF7AIwvjOIYGJQwMkPSigsAPLZhRrAfCe3sgAzID+dDaLEmcNUUBTIL2NQgfLNAkD6k1wg8oqFooUahepCMP00BQC95QvY1zDquQAAAABJRU5ErkJggg==",
"md5": "50170053b55179167f1c21350c869bb7",
"mimeType": "image/png",
"name": "hello.png",
"type": "VISUALIZATION"
}Then we'll "execute" this query on the documentation and get the Blob ID in the response:
{
"id": "0d3fabe0-dc59-406e-820e-fc65a26e0149",
"mimeType": "image/png",
"name": "hello.png",
"type": "VISUALIZATION",
"uploadStatus": "COMPLETE"
}We must make sure to save the Blob ID (0d3fabe0-dc59-406e-820e-fc65a26e0149) as it cannot be retrieved later.
Create an Empty Attachment field
As an example, we're going to use a notebook entry schema with a single Attachment field. See our help documentation for more information on entry schemas. This is just for example purposes though, and the process is nearly identical for all other schematized objects in Benchling.
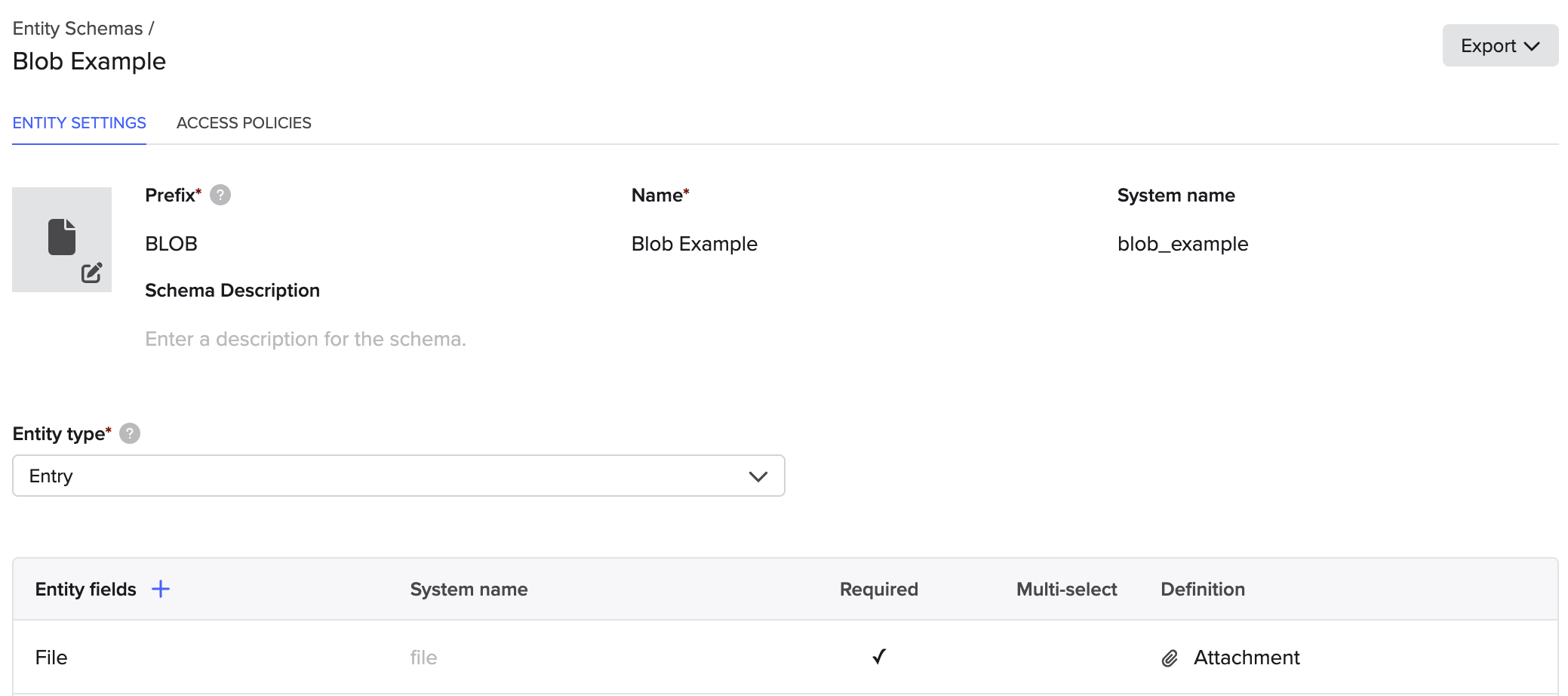
An entry schema configured to have an Attachment field
We'll now create an entry and assign it the example schema.
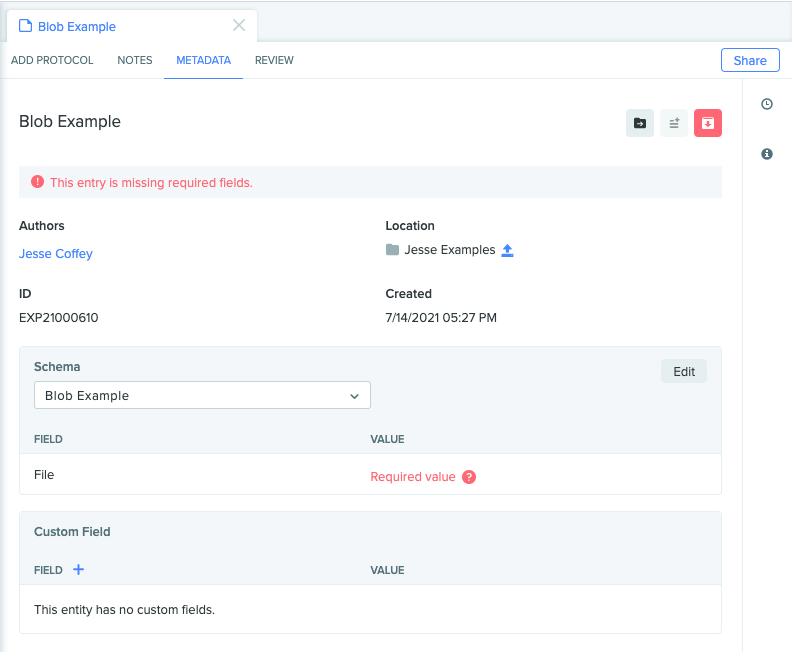
Entry entitled "Blob Example" with the assigned schema and empty Attachment field with name "File".
We now retrieve the API ID of that entry by either:
- Copying the API ID in the UI
- List entries and filter by name
We now have our entry's API ID! For this example, it's etr_ZbYBCbAi.
Now that we have the API ID of the destination parent resource and the BlobID of the file we uploaded, we can finally link the two together.
Link the Blob
To link the blob to the entry, we need to PATCH that field in the entry. To do that, we'll call the PATCH endpoint for entries with the entry ID from the previous step and the proper request body to update the schema field with the blob ID. For example:
Executing it, if successful, it returned the updated entry resource with the new schema field value:
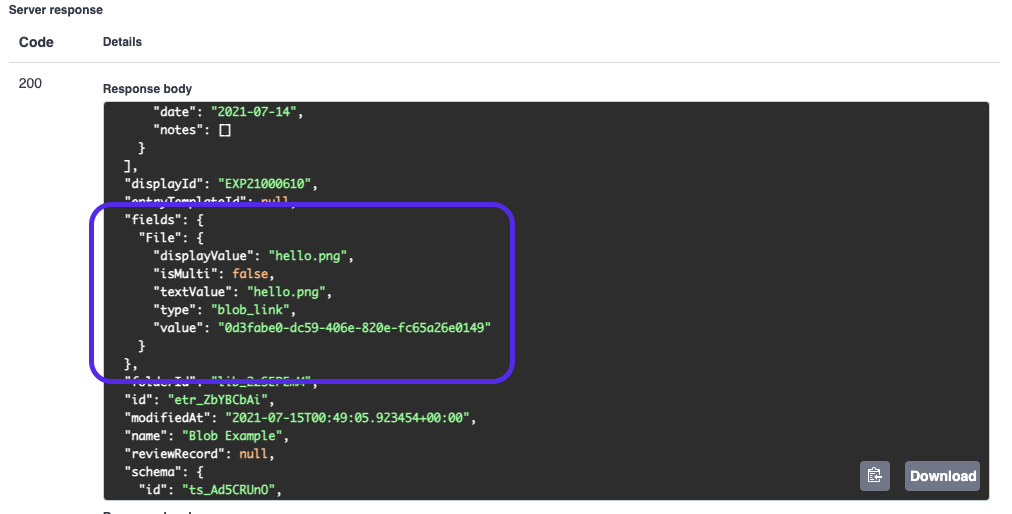
And we can see our file in the UI!
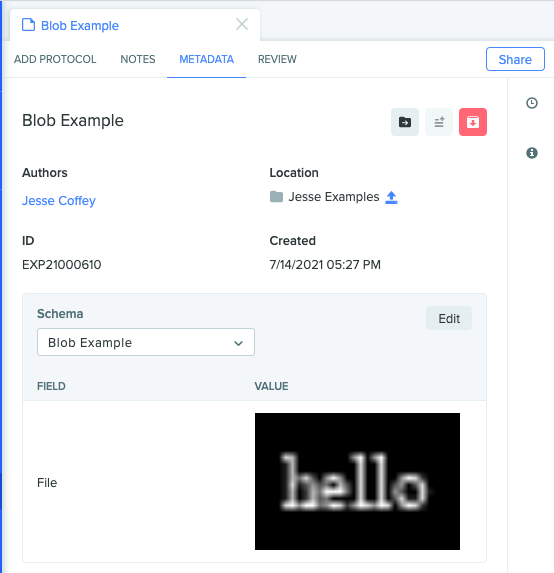
Check out other example for more info on how to use the developer platform, or try this out with new schema types like entities or results!
Updated 7 months ago
