Browsing the SDK Reference Documentation
Benchling SDK Docs are now live!The Benchling SDK documentation site is now live at https://benchling.com/sdk-docs. While the SDK documentation site is the easiest way to view SDK documentation, the content of this guide is still completely up-to-date if you prefer to view the SDK documentation via the methods described below.
Functionality in the SDK is described with Python docstrings. You'll need to inspect these docstrings to access the full reference of what's available in the SDK. They are generally accessed in two ways:
- Python's built-in
help()functionality - Major IDEs support parsing and displaying them, as shown for PyCharm and VSCode below
The SDK is organized into a list of services which map as closely as possible to the Resources list on the Benchling Public API Documentation.
For instance, the API to List Entries is available under the Entries resource category on the lefthand side of the Public API Documentation. In the Benchling SDK, this is done with:
benchling.entries.list_entries()
# ^ - all endpoints are accessed via the Benchling object
# ^ - entries is the category from the API reference
API Support in the SDKWhile the SDK is intended to support all resources in the API, the SDK’s resource support can lag the APIs, so some API resources might not support translation into Python classes. However, they are still accessible to call via the SDK directly.
Using Python's help Functionality
help FunctionalityAfter installing the SDK, you may leverage the help() function to inspect parameter details and documentation of the SDK components.
For example, calling help(benchling_sdk) in a Python shell will reveal the contents of the benchling_sdk package.
Help on package benchling_sdk:
NAME
benchling_sdk
PACKAGE CONTENTS
benchling
errors
helpers (package)
models (package)
services (package)Additionally, you may call help() on any nested object within the SDK. The following line of code:
help(benchling_sdk.benchling)Will return:
Help on module benchling_sdk.benchling in benchling_sdk:
NAME
benchling_sdk.benchling
CLASSES
builtins.object
Benchling
typing_extensions.Protocol(builtins.object)
BenchlingApiClientDecorator
class Benchling(builtins.object)
| Benchling(url: str, auth_method: benchling_api_client.benchling_client.AuthorizationMethod, base_path: Union[str, NoneType] = '/api/v2', retry_strategy: Union[benchling_sdk.helpers.retry_helpers.RetryStrategy, NoneType] = RetryStrategy(max_tries=5, backoff_factor=0.5, status_codes_to_retry=(<HTTPStatus.TOO_MANY_REQUESTS: 429>, <HTTPStatus.BAD_GATEWAY: 502>, <HTTPStatus.SERVICE_UNAVAILABLE: 503>, <HTTPStatus.GATEWAY_TIMEOUT: 504>)), client_decorator: Union[benchling_sdk.benchling.BenchlingApiClientDecorator, NoneType] = None)
|
| A facade for interactions with the Benchling platform.
|
| Methods are organized into namespaces which generally correspond to Resources in Benchling's public API doc.
|
| See https://benchling.com/api/reference
|
| Methods defined here:
|
| __init__(self, url: str, auth_method: benchling_api_client.benchling_client.AuthorizationMethod, base_path: Union[str, NoneType] = '/api/v2', retry_strategy: Union[benchling_sdk.helpers.retry_helpers.RetryStrategy, NoneType] = RetryStrategy(max_tries=5, backoff_factor=0.5, status_codes_to_retry=(<HTTPStatus.TOO_MANY_REQUESTS: 429>, <HTTPStatus.BAD_GATEWAY: 502>, <HTTPStatus.SERVICE_UNAVAILABLE: 503>, <HTTPStatus.GATEWAY_TIMEOUT: 504>)), client_decorator: Union[benchling_sdk.benchling.BenchlingApiClientDecorator, NoneType] = None)
| Initialize Benchling.
|
| :param url: A server URL (host and optional port) including scheme such as https://benchling.com
| :param auth_method: A provider of an HTTP Authorization header for usage with the Benchling API. Pass an
| instance of benchling_sdk.auth.api_key_auth.ApiKeyAuth with a valid Benchling API token for
| authentication and authorization through HTTP Basic Authentication, or a client_id and
| client_secret pair with benchling_sdk.auth.client_credentials_oauth2.ClientCredentialsOAuth2
| for authentication and authorization through OAuth2 client_credentials Bearer token flow.
| :param base_path: If provided, will append to the host. Otherwise, assumes the V2 API. This is
| a workaround until the generated client supports the servers block. See BNCH-15422
| :param retry_strategy: An optional retry strategy for retrying HTTP calls on failure. Setting to None
| will disable retries
| :param client_decorator: An optional function that accepts a BenchlingApiClient configured with
| default settings and mutates its state as desired
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| aa_sequences
| AA Sequences.
|
| AA Sequences are the working units of cells that make everything run (they help make structures, catalyze
| reactions and allow for signaling - a kind of internal cell communication). On Benchling, these are comprised
| of a string of amino acids and collections of other attributes, such as annotations.
|
| See https://benchling.com/api/reference#/AA%20Sequences
|
| api
| Make custom API calls with the underlying BenchlingApiClient.
|
| A common use case for this is making calls to API endpoints which may not yet be supported in the current SDK
| release. It's capable of making more "generic" calls utilizing our authorization scheme, as well as supporting
| some simple serialization and deserialization for custom models.
|
| assay_results
| Assay Results.
|
| Results represent the output of assays that have been performed. You can customize the schemas of results to
| fit your needs. Results can link to runs, batches, and other types.
|
| See https://benchling.com/api/reference#/Assay%20Results
|
| assay_runs
| Assay Runs.
|
| Runs capture the details / parameters of a run that was performed. Results are usually nested under a run.
|
| See https://benchling.com/api/reference#/Assay%20Runs
|
| blobs
| Blobs.
|
| Blobs are opaque files that can be linked to other items in Benchling, like assay runs or results. For example,
| you can upload a blob, then upload an assay result that links to that blob by ID. The blob will then appear as
| part of the assay result in the Benchling web UI.
|
| See https://benchling.com/api/reference#/Blobs
|
| boxes
| Boxes.
|
| Boxes are a structured storage type, consisting of a grid of positions that can each hold one container. Unlike
| locations, there are a maximum number of containers that a box can hold (one per position).
|
| Boxes are all associated with schemas, which define the type of the box (e.g. "10x10 Cryo Box") along with the
| fields that are tracked and the dimensions of the box.
|
| Like all storage, every Box has a barcode that is unique across the registry.
|
| See https://benchling.com/api/reference#/Boxes
|
| client
| Provide access to the underlying generated Benchling Client.
|
| Should generally not be used except for advanced use cases which may not be well supported by the SDK itself.
|
| containers
| Containers.
|
| Containers are the backbone of sample management in Benchling. They represent physical containers, such as
| tubes or wells, that hold quantities of biological samples (represented by the batches inside the container).
| The container itself tracks its total volume, and the concentration of every batch inside of it.
|
| Containers are all associated with schemas, which define the type of the container (e.g. "Tube") along with the
| fields that are tracked.
|
| Like all storage, every container has a barcode that is unique across the registry.
|
| See https://benchling.com/api/reference#/Containers
|
| custom_entities
| Custom Entities.
|
| Benchling supports custom entities for biological entities that are neither sequences or proteins. Custom
| entities must have an entity schema set and can have both schema fields and custom fields.
|
| See https://benchling.com/api/reference#/Custom%20Entities
|
| dna_alignments
| DNA Alignments.
|
| A DNA alignment is a Benchling object representing an alignment of multiple DNA sequences.
|
| See https://benchling.com/api/reference#/DNA%20Alignments
|
| dna_sequences
| DNA Sequences.
|
| DNA sequences are the bread and butter of the Benchling Molecular Biology suite. On Benchling, these are
| comprised of a string of nucleotides and collections of other attributes, such as annotations and primers.
|
| See https://benchling.com/api/reference#/DNA%20Sequences
|
| dropdowns
| Dropdowns.
|
| Dropdowns are registry-wide enums. Use dropdowns to standardize on spelling and naming conventions, especially
| for important metadata like resistance markers.
|
| See https://benchling.com/api/reference#/Dropdowns
|
| entries
| Entries.
|
| Entries are rich text documents that allow you to capture all of your experimental data in one place.
|
| See https://benchling.com/api/reference#/Entries
|
| events
| Events.
|
| The Events system allows external services to subscribe to events that are triggered in Benchling (e.g. plasmid
| registration, request submission, etc).
|
| See https://benchling.com/api/reference#/Events
|
| exports
| Exports.
|
| Export a Notebook Entry.
|
| See https://benchling.com/api/reference#/Exports
|
| folders
| Folders.
|
| Manage folder objects.
|
| See https://benchling.com/api/reference#/Folders
|
| inventory
| Inventory.
|
| Manage inventory wide objects.
|
| See https://benchling.com/api/reference#/Inventory
|
| lab_automation
| Lab Automation.
|
| Lab Automation endpoints support integration with lab instruments, and liquid handlers to create samples or
| results, and capture transfers between containers at scale.
|
| See https://benchling.com/api/reference#/Lab%20Automation
|
| label_templates
| Label Templates.
|
| List label templates.
|
| See https://benchling.com/api/reference#/Label%20Templates
|
| locations
| Locations.
|
| Manage locations objects. Like all storage, every Location has a barcode that is unique across the registry.
|
| See https://benchling.com/api/reference#/Locations
|
| oligos
| Oligos.
|
| Oligos are short linear DNA sequences that can be attached as primers to full DNA sequences. Just like other
| entities, they support schemas, tags, and aliases.
|
| Please migrate to the corresponding DNA Oligos endpoints so that we can support RNA Oligos.
|
| See https://benchling.com/api/reference#/Oligos
|
| plates
| Plates.
|
| Plates are a structured storage type, grids of wells that each function like containers. Plates come in two
| types: a traditional "fixed" type, where the wells cannot move, and a "matrix" type. A matrix plate has similar
| functionality to a box, where the containers inside can be moved around and removed altogether.
|
| Plates are all associated with schemas, which define the type of the plate (e.g. "96 Well Plate") along with
| the fields that are tracked, the dimensions of the plate, and whether or not the plate is a matrix plate or a
| traditional well plate.
|
| Like all storage, every Plate has a barcode that is unique across the registry.
|
| See https://benchling.com/api/reference#/Plates
|
| printers
| Printers.
|
| List printers.
|
| See https://benchling.com/api/reference#/Printers
|
| projects
| Projects.
|
| Manage project objects.
|
| See https://benchling.com/api/reference#/Projects
|
| registry
| Registry.
|
| Manage registry objects.
|
| See https://benchling.com/api/reference#/Registry
|
| requests
| Requests.
|
| Requests allow scientists and teams to collaborate around experimental assays and workflows.
|
| See https://benchling.com/api/reference#/Requests
|
| schemas
| Schemas.
|
| Schemas represent custom configuration of objects in Benchling. See https://docs.benchling.com/docs/schemas in
| our documentation on how Schemas impact our developers
|
| See https://benchling.com/api/reference#/Schemas
|
| tasks
| Tasks.
|
| Endpoints that perform expensive computations launch long-running tasks. These endpoints return the task ID (a
| UUID) in the response body.
|
| After launching a task, periodically invoke the Get a task endpoint with the task UUID (e.g., every 10
| seconds), until the status is no longer RUNNING.
|
| You can access a task for up to 30 minutes after its completion, after which its data will no longer be
| available.
|
| See https://benchling.com/api/reference#/Tasks
|
| warehouse
| Warehouse.
|
| Manage warehouse credentials.
|
| See https://benchling.com/api/reference#/Warehouse
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'_aa_sequence_service': <class 'benchling_sdk.servi...
class BenchlingApiClientDecorator(typing_extensions.Protocol)
| BenchlingApiClientDecorator(*args, **kwds)
|
| For customizing a BenchlingApiClient client, which gives access to the underlying HTTPX layer.
|
| Functions implementing this Protocol will receive the default BenchlingApiClient and may mutate specific
| attributes, returning the updated BenchlingApiClient.
|
| A common use case for this is extending the default HTTP timeout:
|
| def higher_timeout_client(client: BenchlingApiClient) -> BenchlingApiClient: return client.with_timeout(180)
|
| Method resolution order:
| BenchlingApiClientDecorator
| typing_extensions.Protocol
| builtins.object
|
| Methods defined here:
|
| __call__(self, client: benchling_api_client.benchling_client.BenchlingApiClient) -> benchling_api_client.benchling_client.BenchlingApiClient
| Customize a BenchlingApiClient and return the updated client.
|
| :param client: The underlying API client with default configuration
| :type client: BenchlingApiClient
| :return: The updated client
| :rtype: BenchlingApiClient
|
| __init__ = _no_init(self, *args, **kwargs)
|
| __subclasshook__ = _proto_hook(other)
| # Set (or override) the protocol subclass hook.
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __abstractmethods__ = frozenset()
|
| __parameters__ = ()
|
| ----------------------------------------------------------------------
| Class methods inherited from typing_extensions.Protocol:
|
| __class_getitem__(params) from typing_extensions._ProtocolMeta
|
| __init_subclass__(*args, **kwargs) from typing_extensions._ProtocolMeta
| This method is called when a class is subclassed.
|
| The default implementation does nothing. It may be
| overridden to extend subclasses.
|
| ----------------------------------------------------------------------
| Static methods inherited from typing_extensions.Protocol:
|
| __new__(cls, *args, **kwds)
| Create and return a new object. See help(type) for accurate signature.
DATA
Optional = typing.OptionalMethods in the SDK are documented with links to the API Documentation for the operation the SDK method performs. The API documentation may include more detailed information about the behavior of the endpoint or its parameters.
Using IDEs
PyCharm
Ensure your project is using an interpreter where the Benchling SDK is installed.
You may wish to reference: Configure a virtual environment
To peek at documentation for a part of the SDK, you can hover over the class or method name:
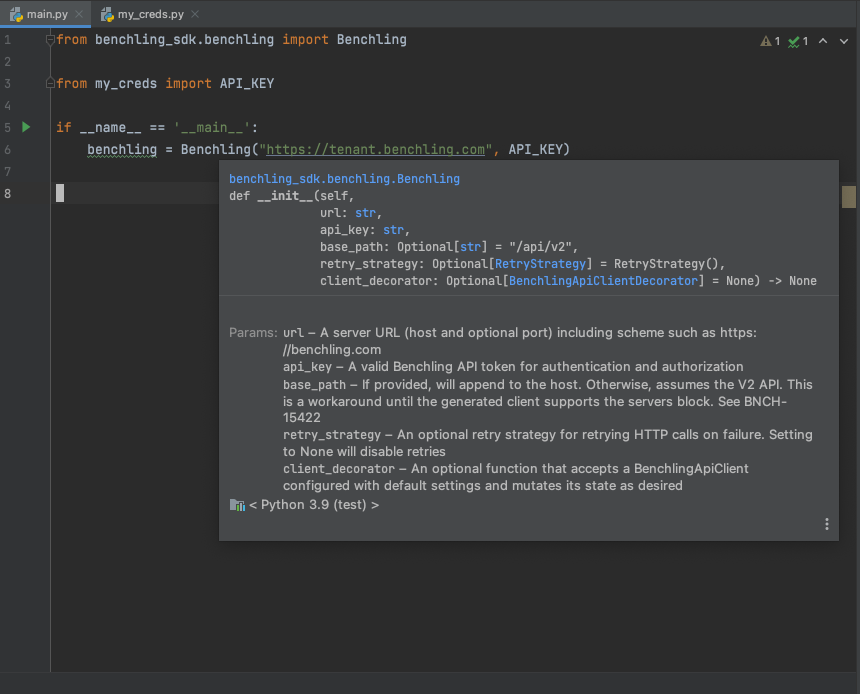
SDK Documentation via PyCharm
To see the documentation within the implementation of the SDK, you can also navigate directly to the source files with Right click → Go To → Declaration or Usages, or Right click → Go To → Implementation(s):
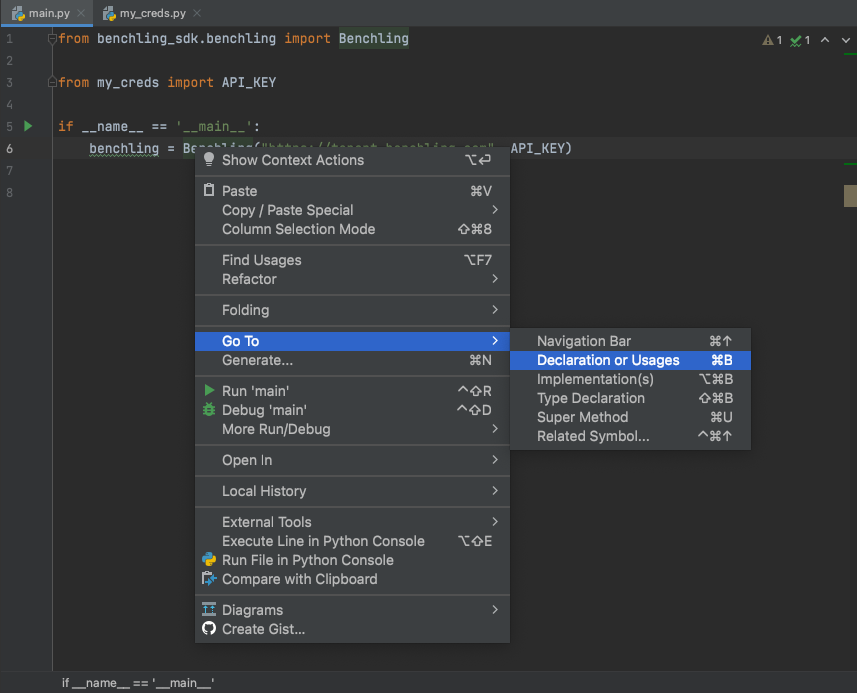
Reveal definitions and declarations of class/method names via PyCharm
VsCode
Ensure your project is using an interpreter where the Benchling SDK is installed. You may wish to reference: Using Python environments in VS Code
To peek at documentation for a part of the SDK, you can hover over the class or method name:
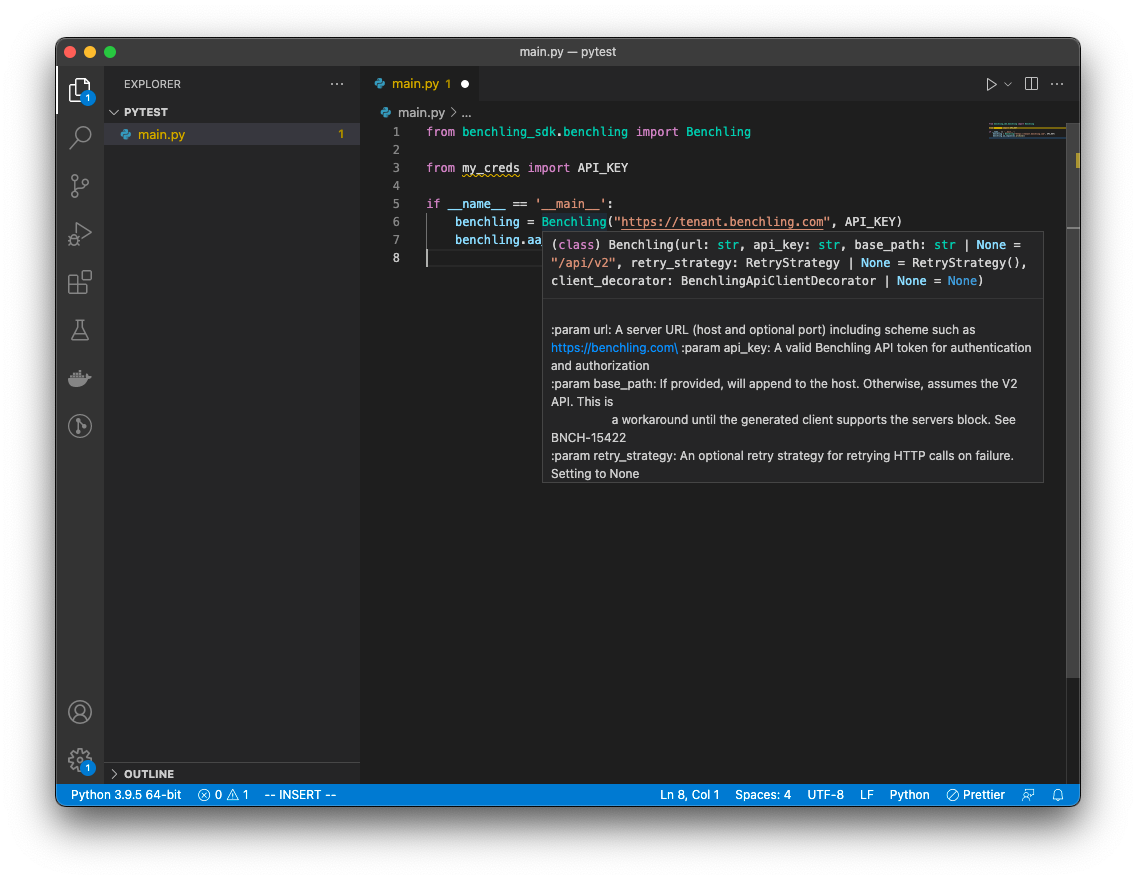
SDK Documentation via VsCode
To see the documentation within the implementation of the SDK, you can also navigate directly to the source files with Right click → Go to Declaration, or Right click → Go to Definition:
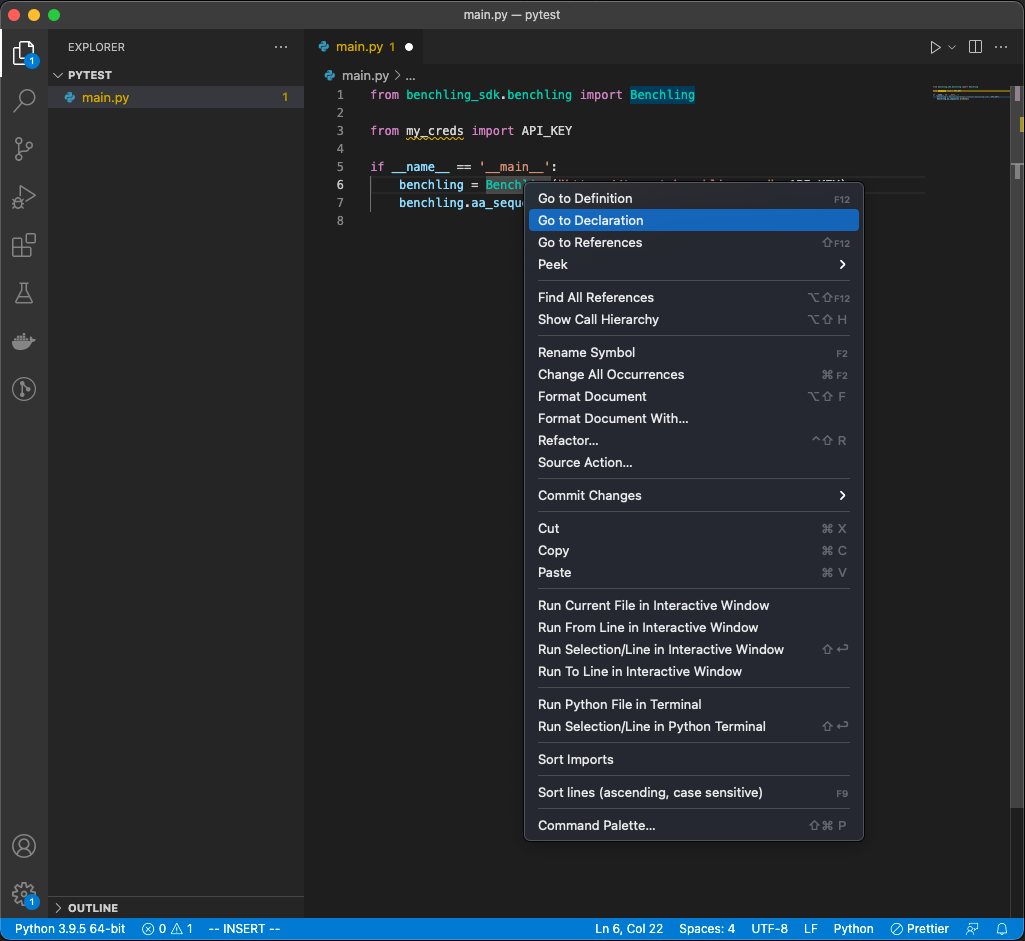
Reveal definitions and declarations of class/method names via VsCode
Updated about 1 year ago
