Overview & Getting started
How to get connected to your secure Warehouse and start exploring your data
Overview
The data warehouse is a read-only SQL database that tracks assay data, biological entities, inventory data, notebook entries, and other records stored in Benchling. The warehouse centralizes the entire organization's research output and facilitates queries that would historically require parsing multiple data sources, such as “find all batches that had an OD > 1000”.
The warehouse is well-suited to complex analytics, where the expressiveness of SQL is important. It can be securely connected to third-party analytics tools such as Tableau and Spotfire to create charts and dashboards backed by Benchling data. Configurable permissions ensure appropriate data access control.
All the data the warehouse stores (and the tables the data are stored in) are outlined in Warehouse Tables .
How it works
The warehouse ingests both user generated data about entities from the registry as well as assay results from networked instruments. To enable this aggregation, the way it works is as follows:
- Researchers register new samples in the registry, specifying properties about the sample. For example, T cell receptor may capture information about the Alpha chain, Beta chain, and Specificity.
- They initiate a new run, specifying the assay to be run, the sample IDs, and parameters such as the adapter used.
- Results from the assay are uploaded as structured data and blobs to the warehouse.
- Third party analysis and visualization tools can connect to the warehouse to consume the data.
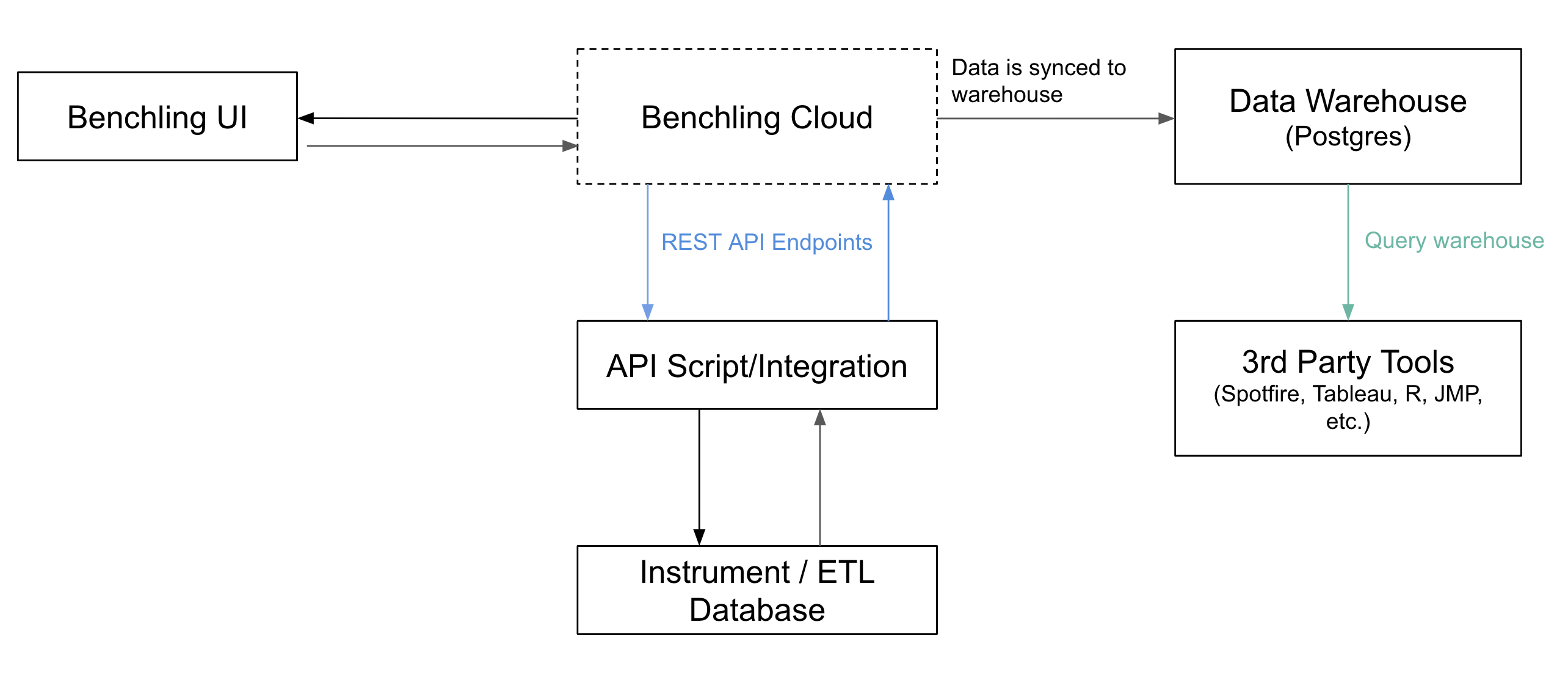
Warehouse architecture
When can I expect data to sync to the warehouse?Generally, data uploaded to Benchling will sync to the data warehouse within 24 hours. Shorter tasks like entity and results uploads with fewer than 1000 rows are often synced to the warehouse within minutes, depending on the total activity in your Benchling tenant. Larger uploads can take longer to sync to the data warehouse, like registry or results uploads with more than 1000 rows and large API integrations.
Don't worry if you don't see your data appear in the warehouse until the next day. If you need your data in real time, consider using the Benchling API to upload or access your data.
Obtaining Credentials
Warehouse credentials are obtained via the Account Settings page in Benchling. From the settings page, you can generate new credentials by clicking on “Create credentials” under the Warehouse credentials section. If you do not see this option, contact your Benchling Customer Success Manager to enable access on your account.
Protecting your Warehouse CredentialsWarehouse credentials can be used to gain direct access to your Benchling data. These credentials are static and can be used by anyone, anywhere, so it's critical to keep them secure. We recommend you keep your warehouse username and password in your favorite password manager or similar secure storage.
Warehouse credentials should not be hard-coded into scripts or checked into source control. If your Warehouse credentials are leaked, they should be immediately revoked and new credentials should be generated via your Profile settings screen in Benchling.
Warehouse Configuration
Benchling Data Warehouse is a hosted PostgreSQL solution. Any client, tool, or system that can connect to a PostgreSQL server can be used to connect to your Warehouse. Connections must be made with SSL/TLS; plaintext (unencrypted) connections are not supported.
What is SSL/TLSTl;dr: SSL/TLS ensures that nobody can read or tamper with your data while it's going over the internet to your Warehouse! You're using it right now to read this document in your web browser, but it can also be used for databases.
Here's a short explanation and a longer one that defines other important SSL/TLS terms.
Security Notice: SSL / TLS and PostgreSQLMany PostgreSQL clients do not by default properly encrypt connections to the database. This includes common tools like
psqlandlibpq-based libraries like Python'spsycopg2. Incorrect configuration puts your data and credentials at risk of an interception attack by a network adversary. Don't assume it works the same as HTTPS in your browser.Our Warehouse server is set to only allow TLS connections (specifically TLS1.2+). But, due to nuances in the PostgreSQL network protocol -- namely the documented but obscure default fallback-to-plaintext behavior -- ultimately security is up to you and the software you connect to your Warehouse.
Please be sure to explicitly enable SSL / TLS in your PostgreSQL client. A correct defense must include root Certificate Authority (CA) verification at the minimum. See below for some common use-cases.
Configuring PostgreSQL Clients for Security
The instructions here set the per-user default and will also work with Python's psycopg2 and the JDBC PostgreSQL Driver in addition to psql. If the instructions below don't work, or you need to connect to many PostgreSQL servers, you will need to refer to the official psql and libpq SSL documentation.
Choosing the Correct Certificate Bundle
The following instructions set up your Warehouse connection to trust the global AWS RDS certificate bundle. (If desired, you can use a more specific RDS Root CA alternative listed here.) These certificates rotate every few years and therefore may need an update if you are getting a connection error.
On Mac OS and Linux:
Set up the ~/.postgresql/root.crt file to have the RDS certificate bundle. Open a terminal and run the following commands (please note this assumes you're using bash as your shell and that you don't already have a root.crt file):
# Download the Aurora RDS root certificate bundle to the default location.
# See https://docs.aws.amazon.com/AmazonRDS/latest/UserGuide/UsingWithRDS.SSL.html for official bundle URLs
curl https://truststore.pki.rds.amazonaws.com/global/global-bundle.pem \
--create-dirs -o ~/.postgresql/root.crt
# Set environment variables to require root CA verification by default
# and make these settings apply to new shells. Use .zshrc for ZSH.
export PGSSLMODE=verify-ca
echo 'export PGSSLMODE=verify-ca' >> ~/.bashrcOn Windows:
Set up the %APPDATA%\postgresql\root.crt file to have the RDS certificate bundle.
- Download the global RDS certificate bundle to your Downloads folder and open that folder. You should see a new file named named, for example,
global-bundle.pem - Open a second windows file explorer window (shortcut: Ctrl+N or go File > New Window) and type
%APPDATA%in its address bar - If you see a folder called
postgresqlhere, great! But if not, create a new folder named exactly that. Open this folder. If you can't see or create this folder, contact your IT team - Drag and drop the downloaded
global-bundle.pemfrom the Downloads folder to the postgresql folder - Rename the
global-bundle.pemfile toroot.crtto set it as the default (recommended). Ifroot.crtalready exists, you can simply appendglobal-bundle.pemto it.
We also highly recommend that you also set the PGSSLMODE environment variable to verify-ca (Win+X > System > Advanced System Settings > Advanced Tab > Environment Variables... button). If you are a Windows IT Administrator with Group Policy access, we highly recommend setting this environment variable and PGSSLROOTCERT to the location of a shared ca certificate file for all users.
Advanced Configuration & Considerations
Secure connections must set at least the verify-ca SSL mode. Either use the PGSSLMODE environment variable, or equivalently the sslmode parameter. Without this, a network attacker could spoof the identity of the Warehouse PostgreSQL server with a fake certificate, and steal everything you have access to.
If you want to set an alternative location for the root CA certificate, you can set the PGSSLROOTCERT environment variable or equivalently the sslrootcert parameter to the path to the file. Note: ~/.postgresql/root.crt (and the equivalent on Windows) can contain multiple certificates if you need to connect and trust multiple PostgreSQL servers; just append the additional PEM-formatted certificates to this file.
The verify-full mode is supported for extra security, but you may need to adjust your connection settings hostname to "pin" it to the actual RDS Aurora PostgreSQL instance. To find the actual RDS PostgresSQL instance name from your "friendly name", e.g. postgres-warehouse.examplename.benchling.com, you can use DNS tools or websites like nslookup.io to find the CNAME for the friendly DNS alias provided by benchling. Be aware that Benchling may need to migrate your data to meet certain performance guarantees and therefore this pinned hostname may change. See AWS's full SSL/TLS configuration advice for Aurora PostgreSQL more info.
Connecting with psql
SSL / TLS and psqlIf you don't put the
sslmodeparameter or have thePGSSLMODEenvironment variable set, psql will default to "prefered" SSL mode, which can be easily hacked via eavesdropping and MITM. Don't get hacked. See above for more details.
To connect to your Warehouse, run (replace yourusername and yourdeployname with your individual settings):
psql 'postgresql://[email protected]:5432/warehouse?sslmode=verify-ca'
# or, equivalently in multiple lines
# (no, psql doesn't have a command-line option for sslmode. ThAnKs PsQl.)
PGSSLMODE=verify-ca psql \
-U 'u$yourusername' \
-h postgres-warehouse.yourdeployname.benchling.com \
-d warehouseThis should prompt for your password. Note: replace yourusername and yourdeployname with your individual settings. Be sure to use single-quotes so the $ doesn't interpolate as a shell variable.
Connect Using JDBC
The connection string for psql will also work for Java JDBC connections: postgresql://[email protected]:5432/warehouse?sslmode=verify-ca
Note: replace yourusername and yourdeployname with your individual settings.
See the PostgreSQL JDBC Connector SslMode documentation for more info on the SSL / TLS settings. You will need to set up the correct RDS root certificates, just like with the other tools above, possibly by using Java's keytool.
Connect Using a Third-Party Tool or SaaS
SSL / TLS and 3rd Party SaaSMany services will require you to enable SSL or TLS to connect -- and it's super important that you do enable it! Please also enable the Root Certificate Authority ("Root CA") verification feature or ask your SaaS vendor to support it.
To connect to the Warehouse, use the following parameters in your data visualization tool:
- Host Name: postgres-warehouse.yourdeployname.benchling.com. Be sure to replace yourdeployname with your enterprise specific domain (that means the part before your enterprise's benchling.com URL)
- Port: 5432 (this is the default)
- Database: warehouse
- Username: the username you generated
- Password: the password you generated
- SSL or TLS mode: Enabled - this is very important for your data security
- Root Certificate Authority / Trusted Certificate: Download / upload the global RDS certificate bundle
If possible, you should use the verify-ca SSL / TLS connection mode (it might be called "check server certificate").
If the tool also asks for a SSL / TLS client certificate, either generate your own to get started.
Warehouse Limits and Best Practices
For integrations that connect directly to the Data Warehouse, Benchling has established limits around concurrent connections and transaction duration. These limits ensure that updates can sync in a timely manner and the Warehouse is able to reliably serve queries.
This includes limits on the number of concurrent connections per-user and per-tenants, as well as guidelines around polling frequency and transaction duration. For more specific details, check out the Warehouse limits and best practices section on our Limits page .
Example
Definitions
Runs store parameters about the assay that will be performed, such as the Instrument ID.
Results capture results generated via the assay which are associated to the samples, such as the Cell Count.
Scenario
Capturing results from a Flow Cytometry run
- Researchers configure schemas for the Flow Cytometer assay in the Benchling UI.
- They set up fields, and specify the data type such as string, float etc. Fields can be links to blobs, which are used for handling raw data or images.
FlowCytometryRun: Run
containerId container_link
instrument text
rawData blob_link
FlowGatingResult: Result
flowRun assay_run_link
CD3+ float
CD4+ float
parentResult assay_result_link- They then initiate the assay on the Flow Cytometer, and upload parameters about the run, such as the Instrument ID, to Benchling. An example run includes:
POST /blobs
Response:
{“blobId”: [“65da6215-a889-49d3-a6da-b5cc0ac60d75”]}
POST /blobs/65da6215-a889-49d3-a6da-b5cc0ac60d75/parts
POST /blobs/65da6215-a889-49d3-a6da-b5cc0ac60d75:complete-upload
POST /assay-runs
Parameters:
[{
“schema”: “FlowCytometryRun”,
“fields”: {“instrument”: “My Instrument”,
“rawData”: “65da6215-a889-49d3-a6da-b5cc0ac60d75”},
}]
Response:
{“assayRuns”: [“9c6da62a-0a9e-4b88-b057-1adabfd31e2b”]}- After the run is complete, a script on the instrument uploads results to Benchling, specifying what sample and container they are associated with, and results such as the CD3+ values. An example of a result looks like:
POST /assay-results
Parameters:
{“assayResults”: [
{
“schemaId”: “assaysch_123456”,
“fields”: {“flowRun”: “9c6da62a-0a9e-4b88-b057-1adabfd31e2b”,
“CD3+”: 0.4, “CD4+”: 0.5}
}
...
]}
Response:
{“assayResults”: [“77af3205-65af-457f-87f5-75462b85075a”, ...]}- The run is attached directly to an ELN entry in Benchling
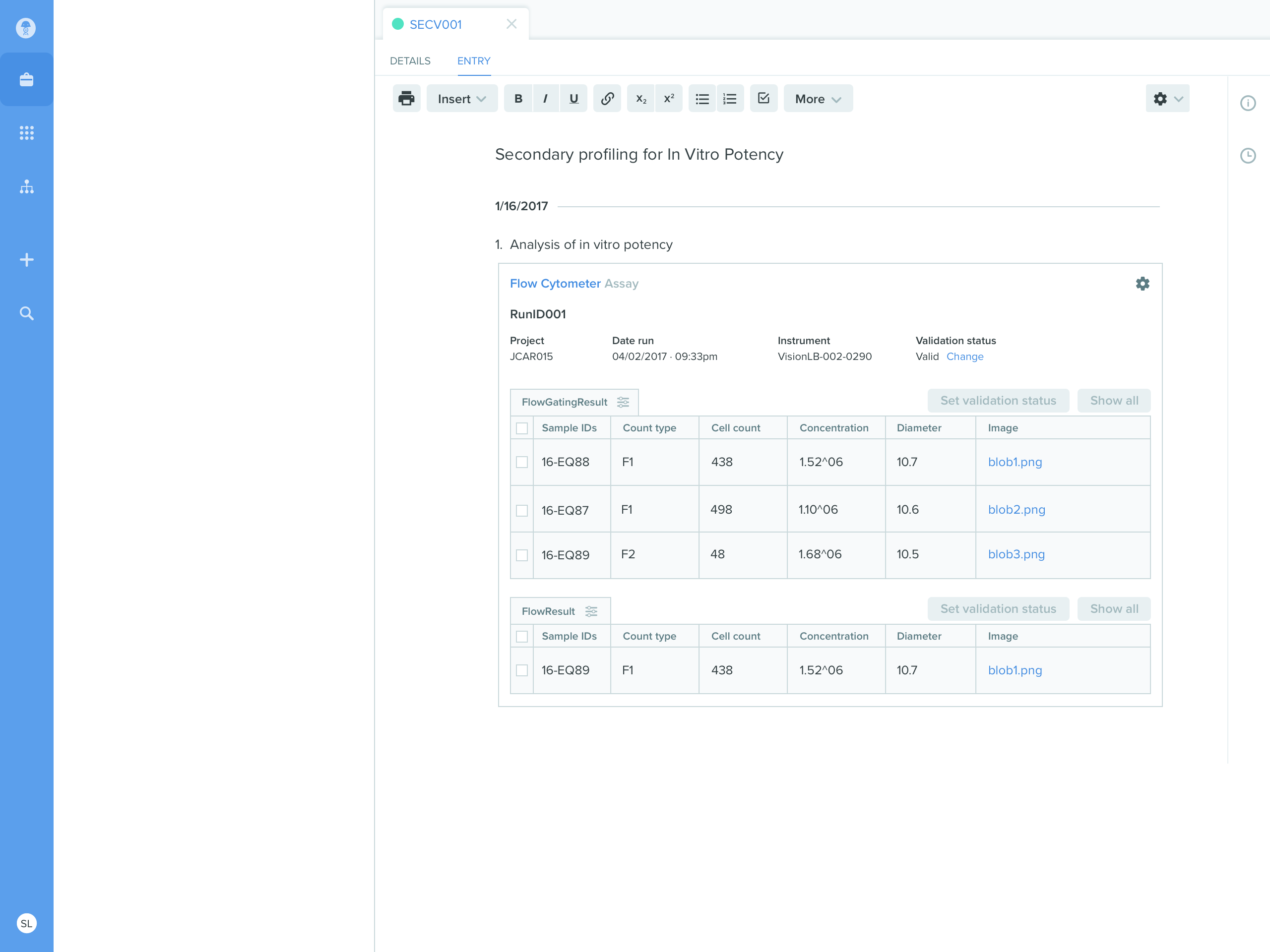
- When researchers want to analyze results across multiple runs, they query the warehouse using either third party analytics tools or through SQL queries
$ psql -h postgres-warehouse.biotechtx.benchling.com
-- Get all batches with CD3plus > 0.5
SELECT batch.id FROM batch
JOIN container ON container.batch_id = batch.id
JOIN flow_cytometry_run ON flow_cytometry_run.container_id = container.id
JOIN flow_gating_result
ON flow_gating_result.flow_run = flow_cytometry_run.id
WHERE flow_gating_result.CD3plus > 0.5
AND flow_gating_result.created_at > ‘2017-01-01’;Updated 7 months ago
