Overview & Tutorial
Overview
The Benchling API is the most flexible way to build an integration with Benchling. The API provides CRUD (Create Read Update Delete) access to almost all data in Benchling, through REST endpoints that represent that data as JSON.
Take a look at our Tutorial to get started with using the API. This will walk you through getting API access, and making some basic calls.
The reference docs contain a list of all the endpoints we offer, as well as examples for how to call each of them.
Building an app or integration?If you're looking to create a persistent application on Benchling that integrates Benchling with another system, you should also check out Getting Started with Benchling Apps, as that is the way we recommend developers build long-running applications on the Developer Platform rather than one-off scripts.
The Python SDK
Benchling has a Python SDK that provides help functions and abstractions for interacting with the APIs! If you're using Python, we highly recommend using it (rather than rebuilding it yourself with the requests package). Check it out here Getting Started with the SDK. The SDK works with either a user API Key or an app's OAuth credentials, so it can be used any time you're calling the APIs in Python.
Tutorial
Welcome to the Benchling API! In this tutorial, we’ll walk you through how to access the API and make your first API call to list the DNA sequences you’ve created in Benchling.
Setup
First, we need to get access to the Developer Platform.
If you already have Developer Platform access, you'll need to generate a personal user API key on your user settings page:
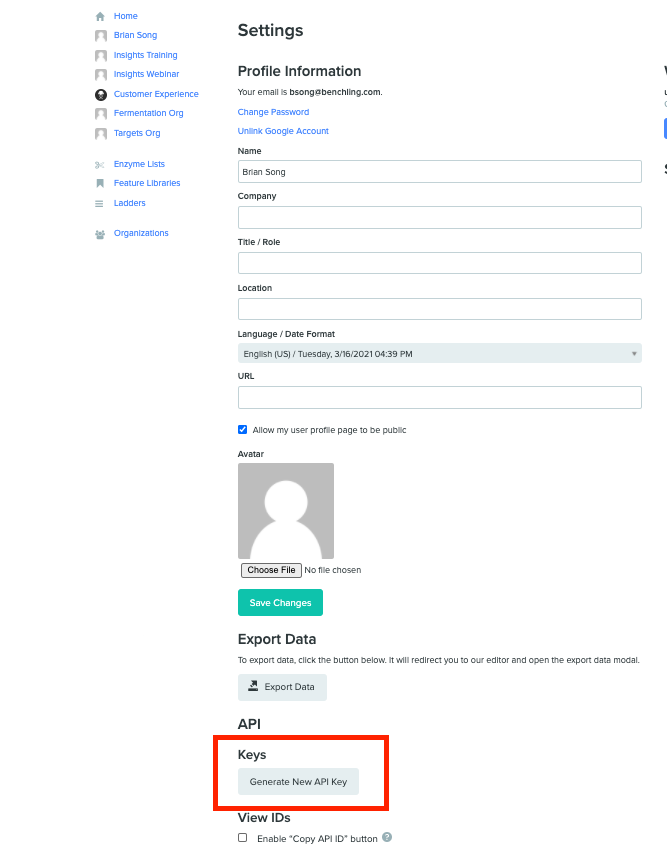
This will allow you to make calls to the API as yourself and allow you to use the interactive API docs at {your-tenant}.benchling.com/api/reference.
The API and Benchling Domains
Your API key is unique for your Benchling domain (sometimes also referred to as your Benchling tenant). When making calls to the API your API key will only be valid for your specific Benchling domain. For example, if your Benchling instance is example.benchling.com (https://example.benchling.com/) and your API key is sk_12345, an example like
curl $YOUR_DOMAIN -u $YOUR_API_KEY:needs to be replaced with
curl https://example.benchling.com -u sk_12345:before you run the command.
List all DNA sequences
First, we'll walk you through listing all the DNA sequences you have access to in Benchling using the List DNA Sequences endpoint.
To explore the API or test out specific endpoints, you can do so directly from the documentation on your domain. Each benchling domain has a version of the API documentation available at {your-domain}.benchling.com/api/reference (e.g. demo.benchling.com/api/reference). The difference is that if you have a login and active API key, you can make calls from within the documentation against the data on that domain.
The public version of the API reference documentation resides at benchling.com/api/reference. This is used as a reference for the API endpoints and their functionality, but cannot be used to make calls to the API.
Calling the API through the documentation produces the same output as calling the API via cURL locally or in your language of choice, and can often be very helpful when trying to debug or understand more about how the API functions.
To make calls, navigate to the endpoint you want to test in the documentation on your domain. We'll use demo.benchling.com for this example, but remember you'll need to visit the domain where you have API access.
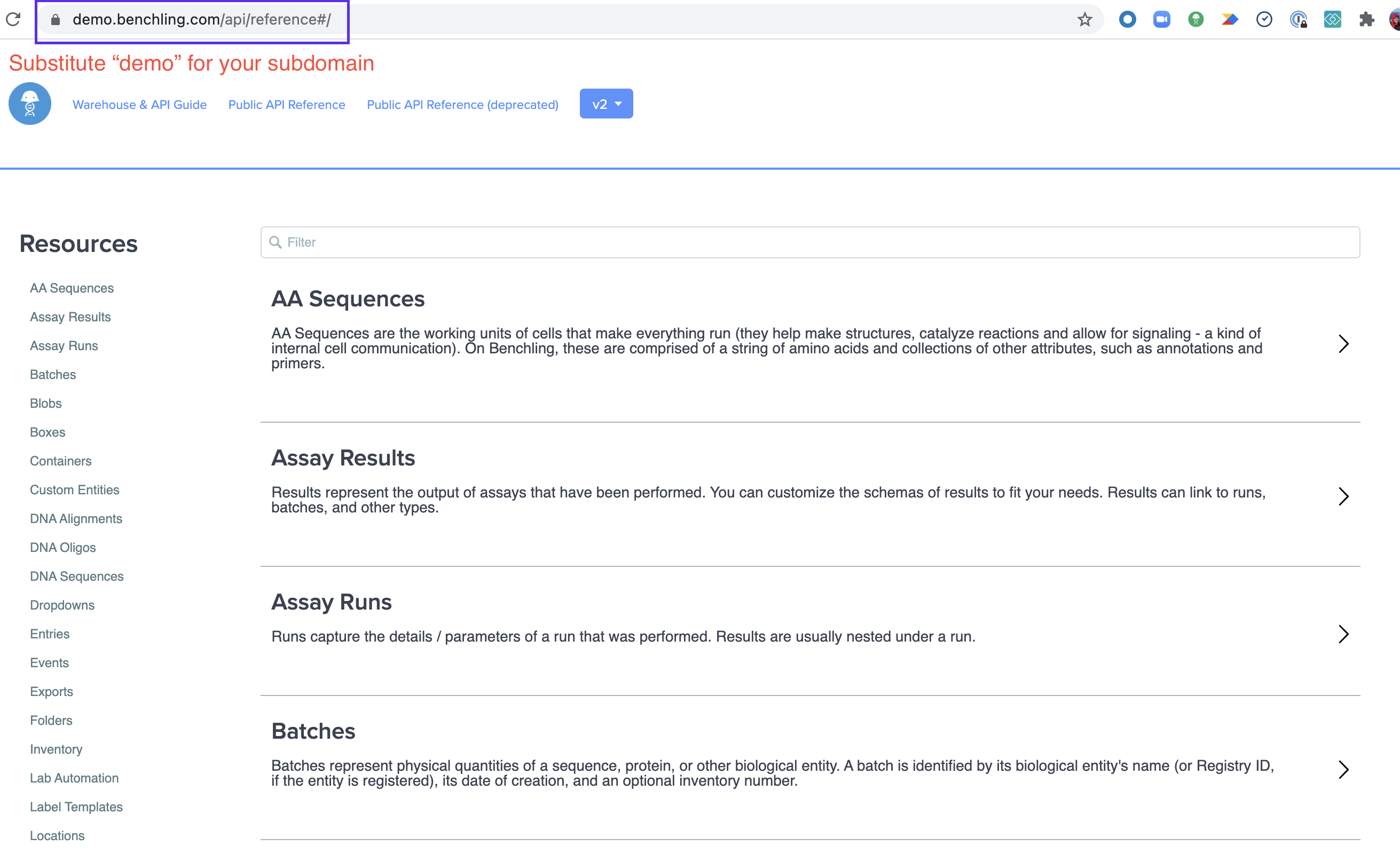
Once there, find the endpoint you'd like to query.
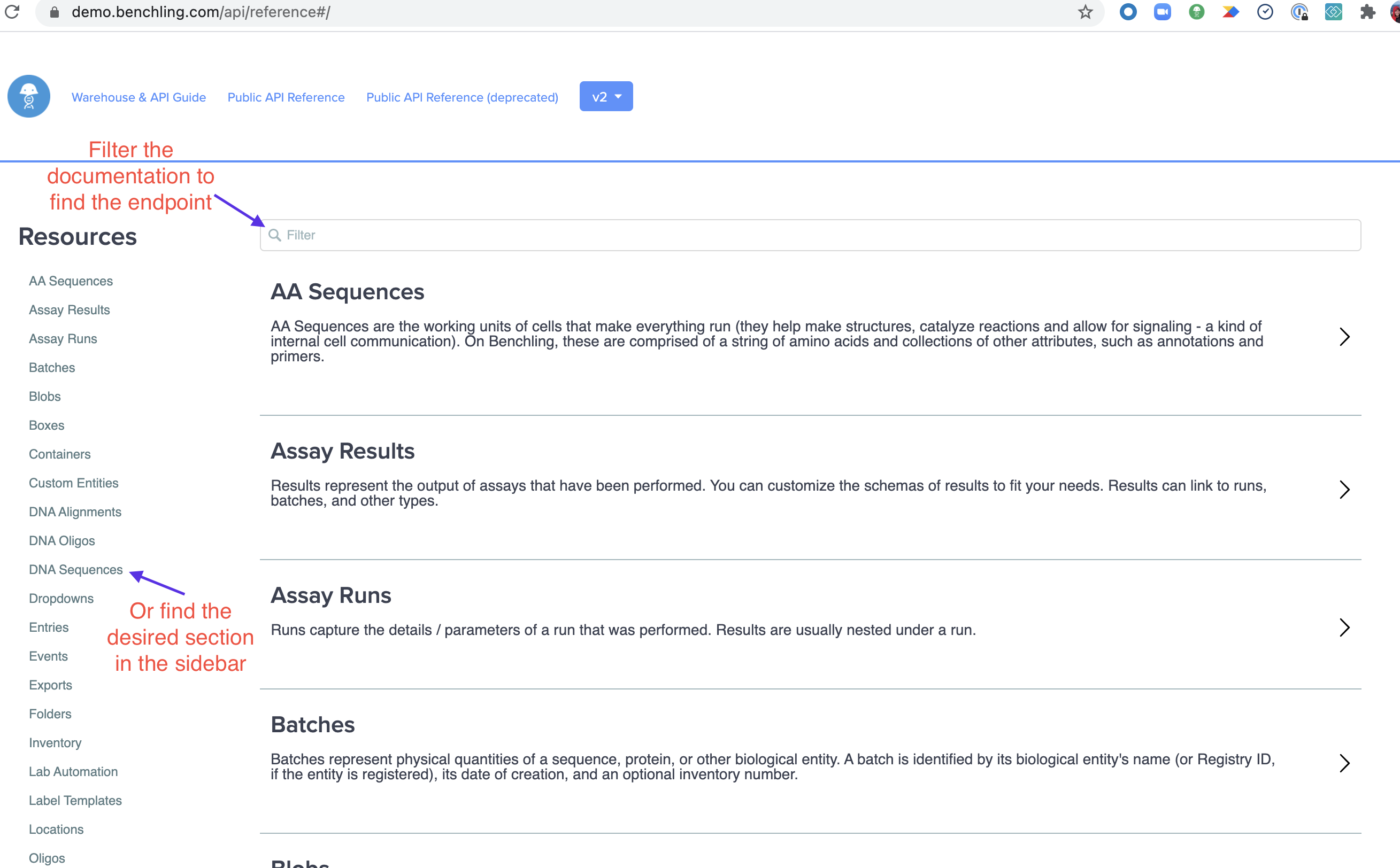
Select the sections to expand them, and select the endpoint to expand its details. Again, this is the List DNA Sequences endpoint at {your-domain}.benchling.com/api/reference#/DNA%20Sequences/listDNASequences. Once expanded, you can start to construct your query by pressing the "Test API" button.
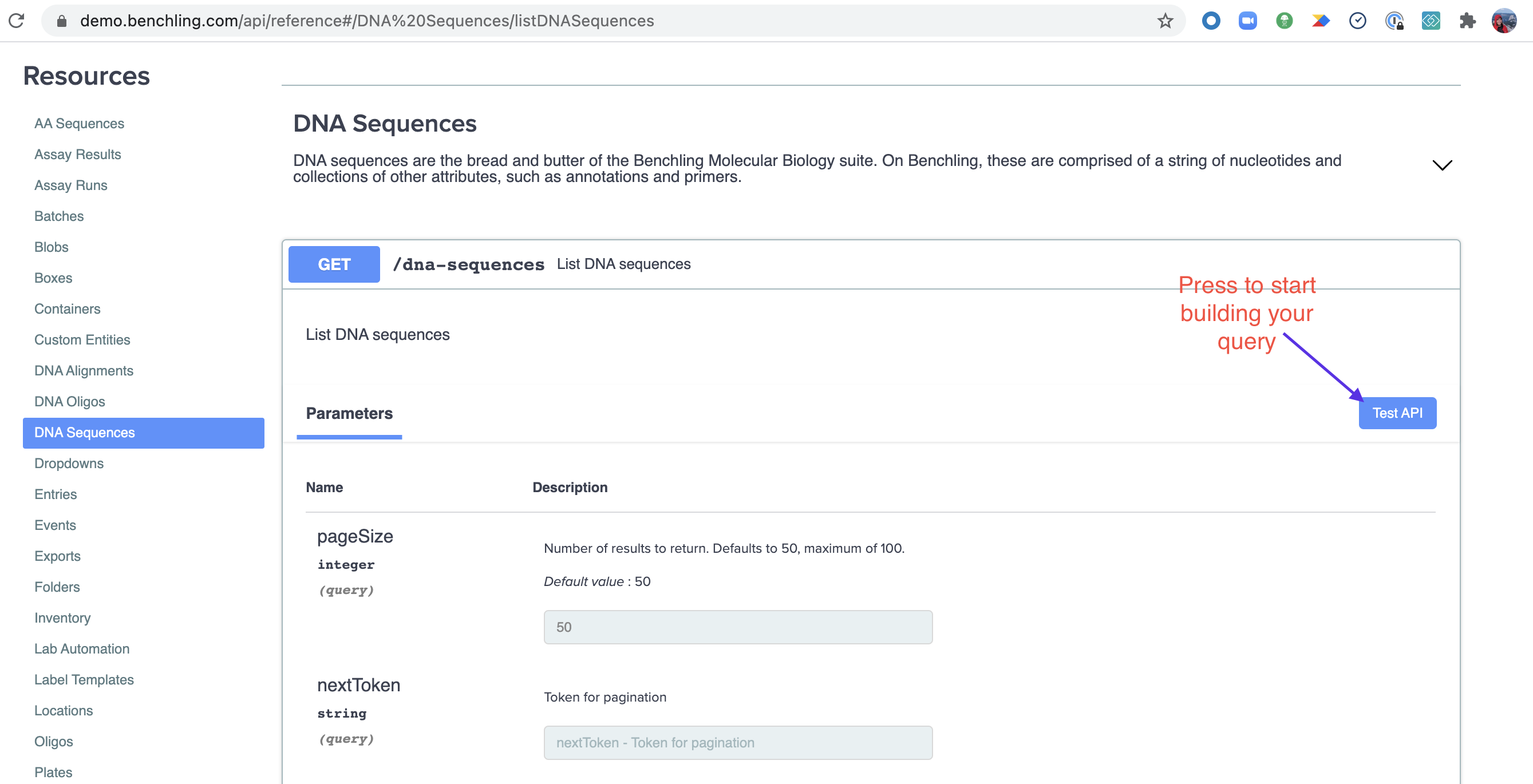
Now you can start filling out your query parameters. We'll leave the defaults for this example since we want to list all the sequences you have access to (up to 50 per query due to pagination).
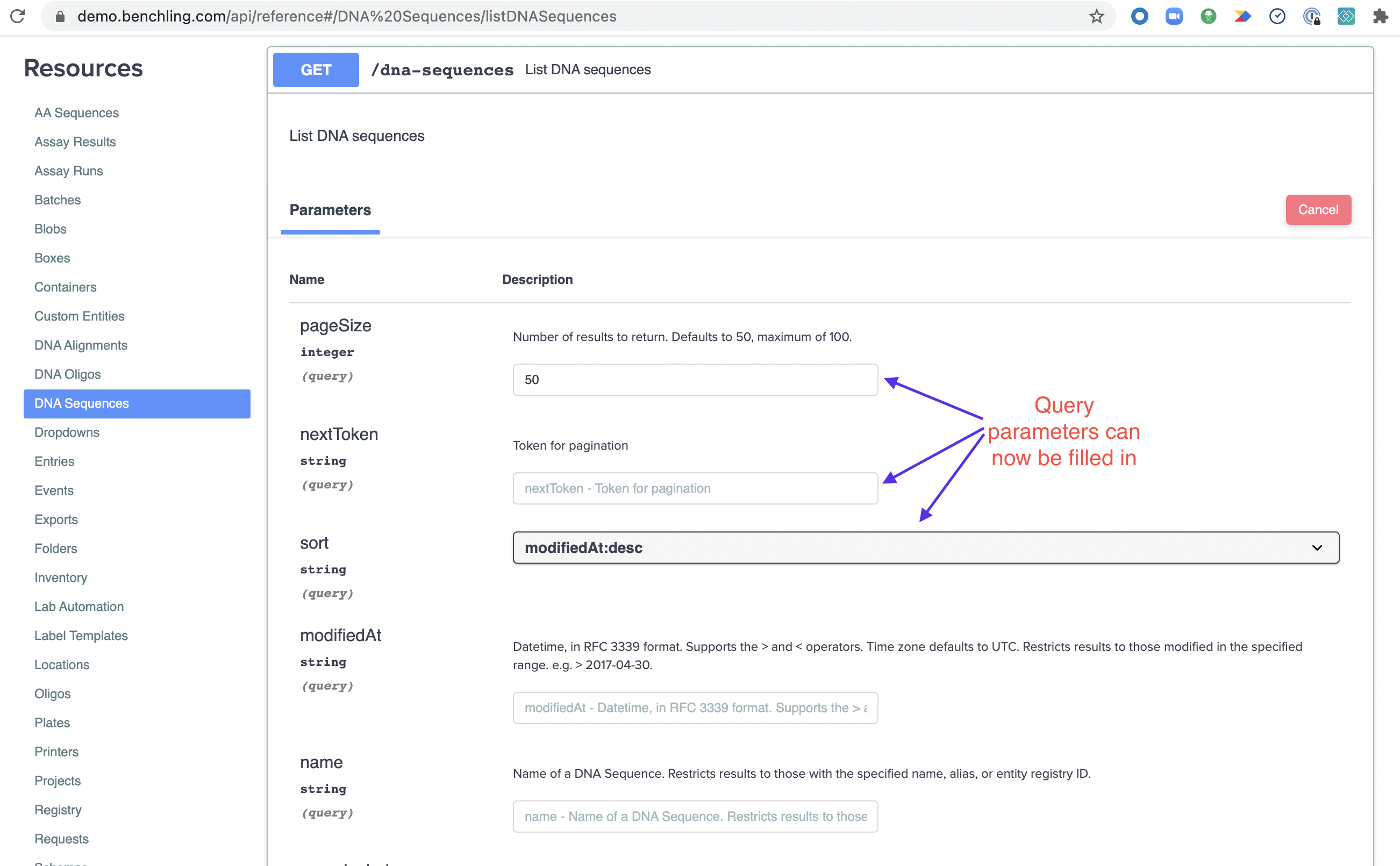
Once you've constructed your query, scroll down and hit "Execute" to have it run the query and call the API.
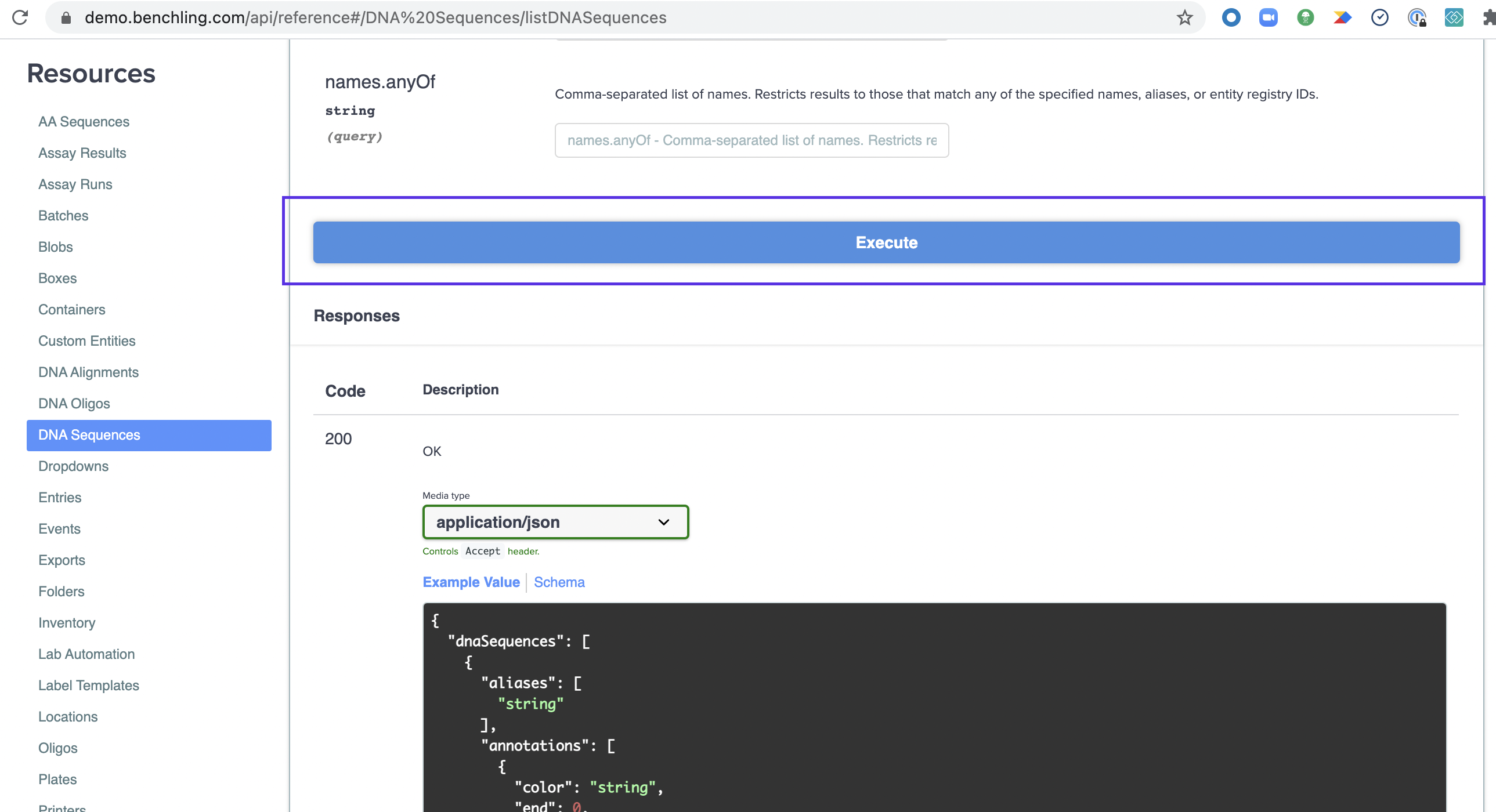
Once it finishes running, you can view the response below in formatted JSON. This is live data from your tenant, the same as would be returned by running it with cURL or the language / tool of your choice. You can test this by copying the command in the "Curl" section and running it on your command line. It automatically generates this when you execute the query, including your credentials (so don't share it!).
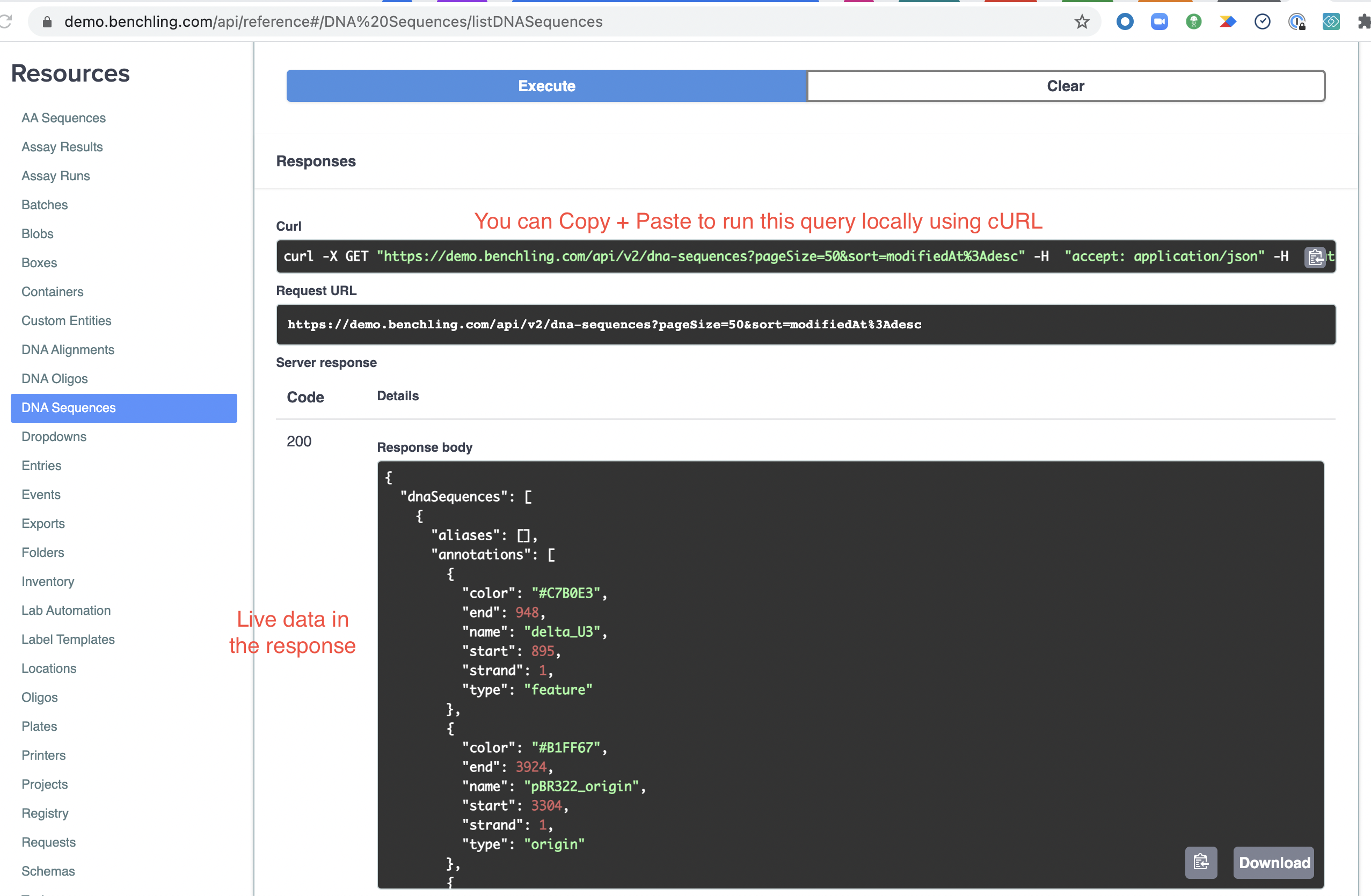
If the query doesn't return any data, you might need to go create a sequence in Benchling's UI (or the API) to have data to retrieve.
List all DNA sequences in a projects
Now we'll show you how to use some of the filter parameters using cURL. The List DNA Sequences endpoint has several options for filtering, and one way to filter the results is to only list the DNA sequences in a particular Benchling project. Filtering by project requires multiple steps:
- First, get the ID of your project by using the List Projects endpoint, or by copying it directly from Benchling (see Copy API IDs).
- Then, pass the project ID to the List DNA Sequences endpoint.
First, call the List Projects endpoint:
curl https://$YOUR_DOMAIN/api/v2/projects -u $YOUR_API_KEY:The endpoint returns a JSON object with an array of projects in the projects field (see the "Example Value" in the documentation). Each Project resource will have an id field with an identifier like src_muzHDKze.
Then, pass the project ID to the List DNA Sequences endpoint, replacing $YOUR_PROJECT_ID with the ID you got from the List Projects call.
curl https://$YOUR_DOMAIN/api/v2/dna-sequences?projectId=$YOUR_PROJECT_ID -u $YOUR_API_KEY:List registered DNA sequences
If you are using the Registry application, you can use the Benchling API to list all the DNA sequences in your registry that belong to a particular schema. First, make sure that you’ve configured a schema and created and registered a DNA sequence. Then, to filter for registered DNA sequences of that schema, you can pass in two extra parameters to the List DNA Sequences endpoint:
registryId, the ID of your registry in Benchling.schemaId, the ID of your schema.
First, find the ID of your registry using the List Registries endpoint:
curl https://$YOUR_DOMAIN/api/v2/registries -u $YOUR_API_KEY:The endpoint will return a list of Registries (see "Example Value" in the documentation), and each Registry resource will have an id field with an identifier like src_gfNcj1gd. If there are multiple registries, you can find the one that’s owned by your organization by looking at the owner field.
Then, find your schema’s ID using the List Entity Schemas endpoint, or by copying it directly from Benchling (see Copy API IDs). Run the following command, replacing $YOUR_REGISTRY_ID with the registry ID you obtained from the List Registries endpoint:
curl https://$YOUR_DOMAIN/api/v2/registries/$YOUR_REGISTRY_ID/entity-schemas -u $YOUR_API_KEY:The endpoint will return a list of Entity Schemas (see "Example Value" in the documentation). To find your schema’s ID, find the schema with the name you are looking for, and then find the id field. The id field will have an identifier like ts_EM122lfJ.
Finally, we’re ready to list DNA sequences. Call the List DNA Sequences endpoint using the registry ID ($YOUR_REGISTRY_ID) and schema ID ($YOUR_SCHEMA_ID):
curl https://$YOUR_DOMAIN/api/v2/dna-sequences?registryId=$YOUR_REGISTRY_ID&schemaId=$YOUR_SCHEMA_ID -u $YOUR_API_KEY:Diving Deeper into API Resource Descriptions
The objects our endpoints return can be complex and benefit from description at a per-field level. We have those types of descriptions and typing information for all of the fields and nested resources available in the "Schema" view:
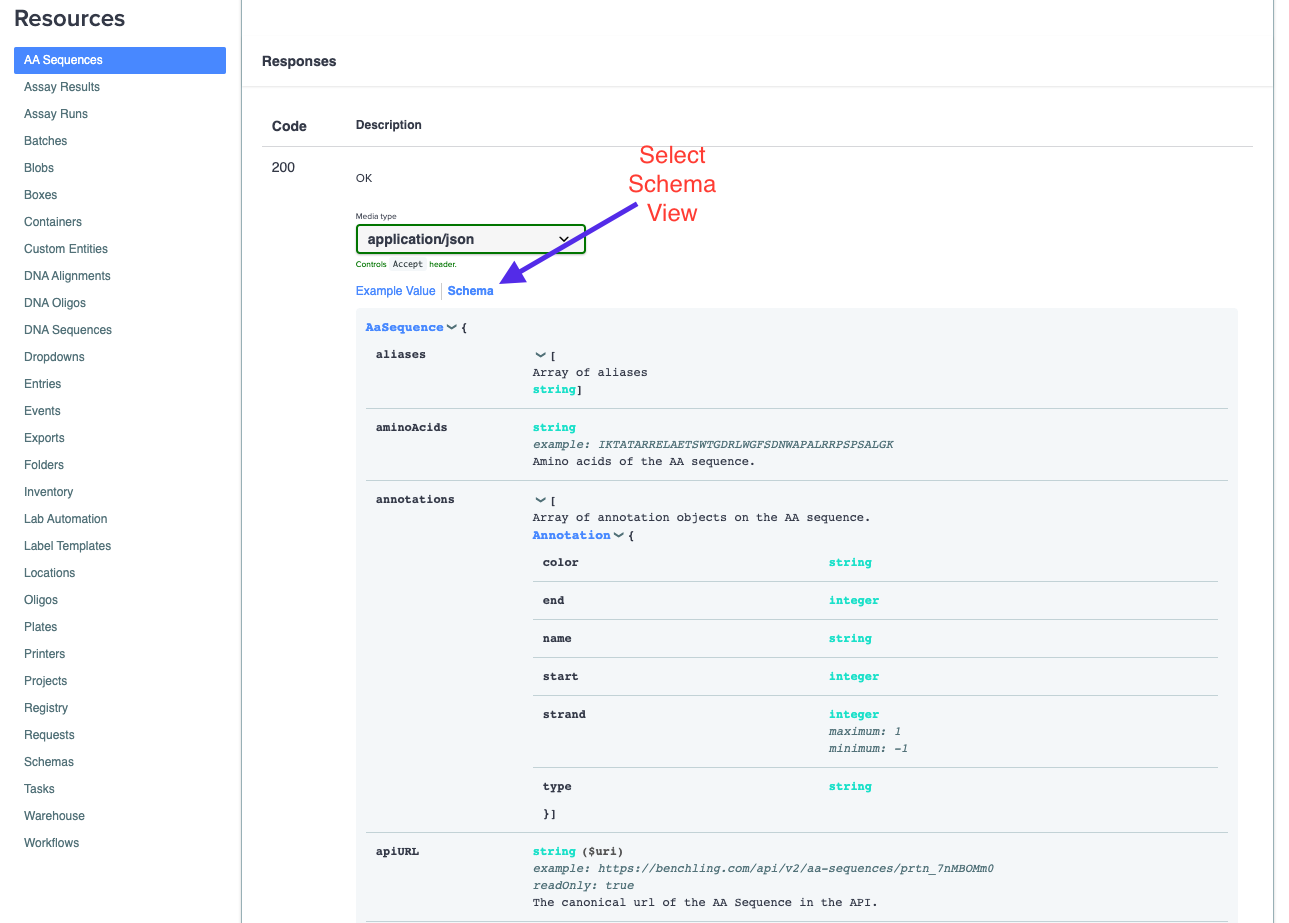
This schema view shows all of the type information and descriptions for all the top level fields that are in the JSON returned by the given endpoint. The above example is the GET AA Sequence endpoint. You can also dive deeper into the nested fields and resources in the response by expanding the respective fields in the schema:
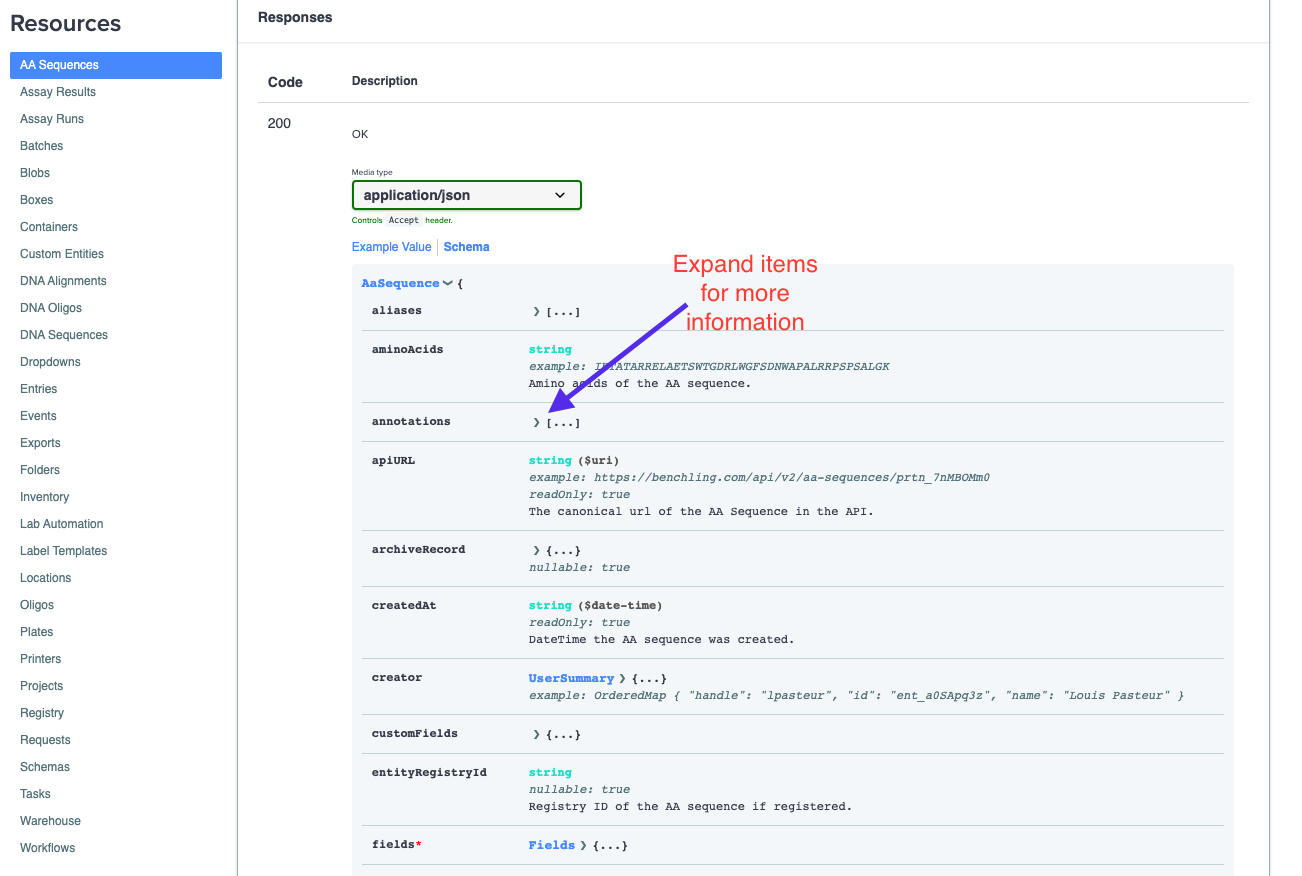
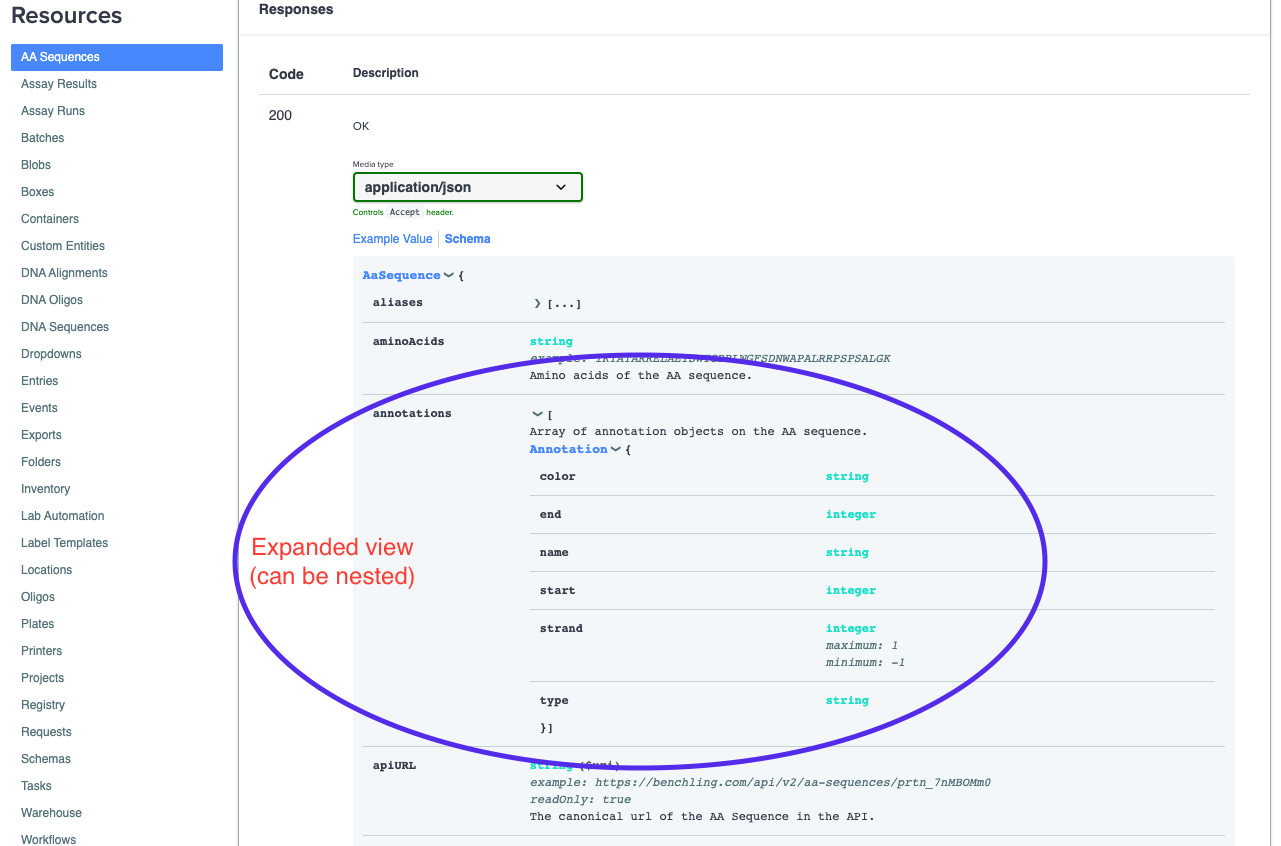
Many of Benchling's objects can have several layers of nesting, so dive deep whenever you're looking for detailed information on a specific field!
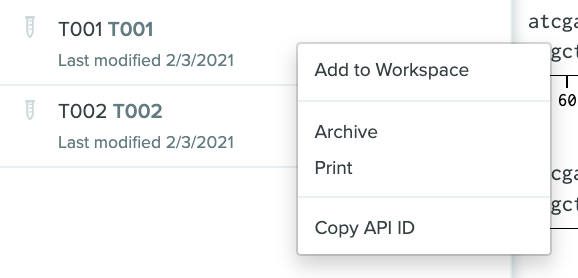
Copy API IDs
Benchling provides a user setting that makes it easy to copy API IDs from the site. To enable it, navigate to the User Settings page, and you'll find the setting in the API section.
Once you've enabled it, you will be able to copy the API ID of schemas, schema fields, dropdowns, or dropdown options on the page where they are listed:
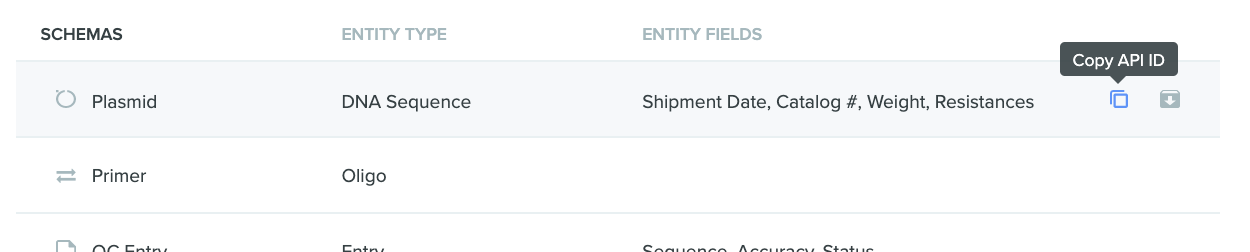
Similarly, you can copy the API ID of entities, inventory, entries, folders, and projects by right-clicking on the row where they are listed.
Returning Query Parameter
The returning parameter is currently implemented on many of the bulk listing endpoints including plates, entries, and entities. The parameter allows users to specify a comma-separated list of fields to return and will modify the output shape to only include the specified fields. To return all keys at a given level, enumerate them or use the wildcard "*".
curl https://$YOUR_DOMAIN/api/v2/plates?returning=plates.id,plates.webURL,plates.creator.name -u $YOUR_API_KEY:Responds only with the id, webURL, and creator's name for each plate object returned as specified by the returning parameter.
{
"plates": [
{
"creator": {
"name": "Louis Pasteur"
},
"id": "plt_VOPAHvfa",
"webURL": "https://local.bnch.us:5000/samples/plates/plt_VOPAHvfa"
},
{
"creator": {
"name": "Louis Pasteur"
},
"id": "plt_VdPMNt5q",
"webURL": "https://local.bnch.us:5000/samples/plates/plt_VdPMNt5q"
}
]
}curl https://$YOUR_DOMAIN/api/v2/plates?returning=plates.id,plates.wells.*.id -u $YOUR_API_KEY:Try this out yourself to see the response!
More examples
For more examples of how to use the Benchling API, you can take a look at Benchling's public repository of example integrations or explore more endpoints in the documentation using the same process we stepped through to list DNA sequences.
IDs in the API
There are a few types of identifiers used in Benchling:
- ID
- Registry ID
- Name
- Alias
For developer use cases, we generally recommend using an objects ID (also referenced as API ID or UUID), as appropriate for the type of object you're working with. These are guaranteed to be stable identifiers on the tenant.
Registry IDs, Names, and Aliases can often be stable enough for many use cases, but are user-editable or programmatically updated under certain circumstances. We recommend using them with caution as appropriate for your project. See our help documentation for more information on how Registry ID, Name, and Alias can be configured.
Note: All identifiers are scoped to the tenant only, and can conflict across tenants. If you require a universally unique ID when operating across more than one Benchling tenant, you should combine ID with the tenant name.
Updated about 1 year ago
