Extending Benchling for NGS Teams
Introduction
Many Benchling customers use Next Generation Sequencing (NGS) for a variety of applications, from sequence confirmations to variant detection. NGS data is not only large and complex, but it is also integral to many of the scientific processes that our customers implement today. Using Benchling Connect and Benchling’s Developer Platform, Benchling customers can integrate data from their NGS instruments with their Benchling applications. This integration gives end users and scientists a smooth experience, allowing them to interact with critical data with the least amount of effort.
What you'll need:
- Access to Benchling Connect
- Access to the Benchling API
- A Benchling App
Definitions
Terms & Concepts
| Term | Definition |
|---|---|
| Sequence Barcode / Index Adapter | Oligo sequences that are ligated at one or both ends of the sample sequence during Library Prep. These sequences are distinct per-library, and are trimmed from the sequence of interest early in secondary analysis |
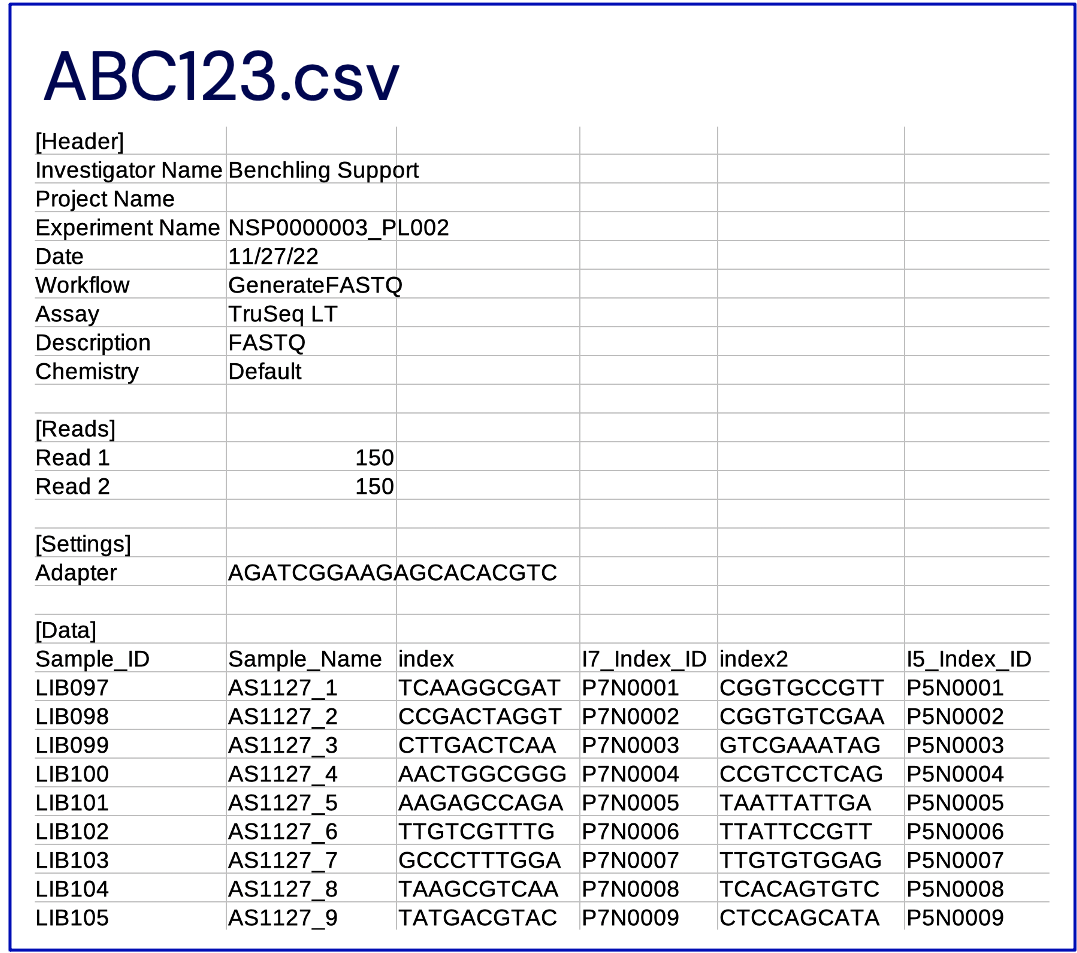
| SampleSheet | NGS SampleSheets are flat files (i.e. CSV-like) that can be passed to the NGS processing pipeline to facilitate demultiplexing and allow for data provenance from primary analysis to results capture back in Benchling. NGS SampleSheets have two main sections: the header section contains metadata about the NGS run and run parameters, while the data section (denoted with the [data] header) contains sample-level information about the samples, IDs and related index barcodes for every entity going through NGS processing. |
| Demultiplex | The process of separating sequences into different FASTQ files per-Library, for all libraries in a pooled sequencing run. There are many tools that can perform this action, like “bcl2fastq” or “demuxfastqs” |
| Primary and Secondary Analyses | Primary analysis assesses raw sequencing data for quality and is commonly performed by software built into the sequencer. Secondary analysis converts data to results, such as alignment and expression, with the use of several bioinformatic tools. |
Scope
NGS Integrations are often designed with three main objectives in mind:
- Sample Sheet Generation
- Instrument run and analytical pipeline monitoring
- Automated upload of secondary analysis results back to Benchling
Sample Sheet Generation
Today, customers often generate SampleSheets manually or through a disconnected, purpose-built software. While this solution works, it takes more time and knowledge from the end scientists than it should. At best, this samplesheet should be generated from a platform that is aware of the entity identifiers and index barcodes, like Benchling.
Instrument run and analytical pipeline monitoring
As the NGS execution and analysis pipeline progresses, we may keep the scientist aware of the status of this pipeline, which can take many hours. This could provide the scientist with valuable information about the error conditions of the pipeline, and help them estimate the health and completion of the pipeline, and reduce the amount of time the end user spends checking up on their runs.
Automated upload of secondary analysis results back to Benchling
Upon completion of the NGS analysis pipeline, the results may be recorded against relevant entities back in a Benchling Notebook entry. In a desired state, the integration will call the Benchling APIs to upload files and results data against schematized results tables. In the UI, the user will view the updated results data or analysis status.
Out of Scope
Benchling is best equipped to handle the entity metadata and results related to NGS experiments. Raw sequencing data (e.g. .bcls, FASTQ) should not be uploaded back into Benchling as they require extra processing (i.e. secondary analyses) in order to derive conclusions from the data.
Keep in mind that these files, though they won't be stored in Benchling, are still important for the provenance of the entities stored in Benchling: It's best practice to consider how the files may still be linked to Benchling entities. For example, a link to the FASTQ file location may be stored as a result on a Library entity in Benchling.
High Level Integration Details
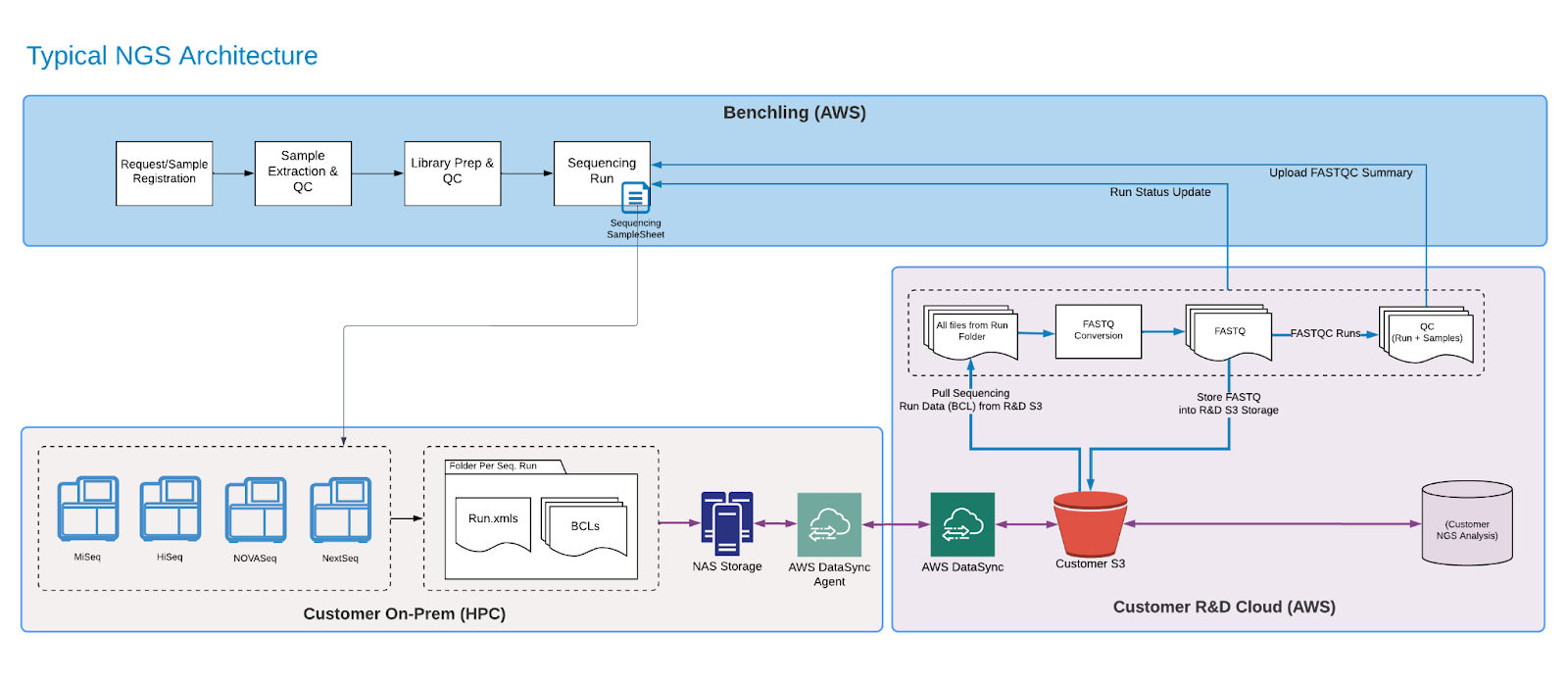
Using the Benchling Developer Platform, developers can establish an integration between Benchling and their NGS technical pipeline. Use cases can span from SampleSheet Generation to results capture.
Integrating with Benchling Connect
Benchling Connect's Runs provide many features that make sense as an integration target for NGS integrations. Runs are templatized, executable from within a notebook entry, and provide a target to push updates to for both automation inputs and outputs via Benchling's REST API. Samplesheet generation, instrument monitoring and results capture can all occur within one object, at one place in a notebook entry that already corresponds to an experiment execution. Below is an example of a user flow for an NGS integration driven by Benchling Connect, using cloud services and a High Performance Computing (HPC) environment as integration touchpoints:
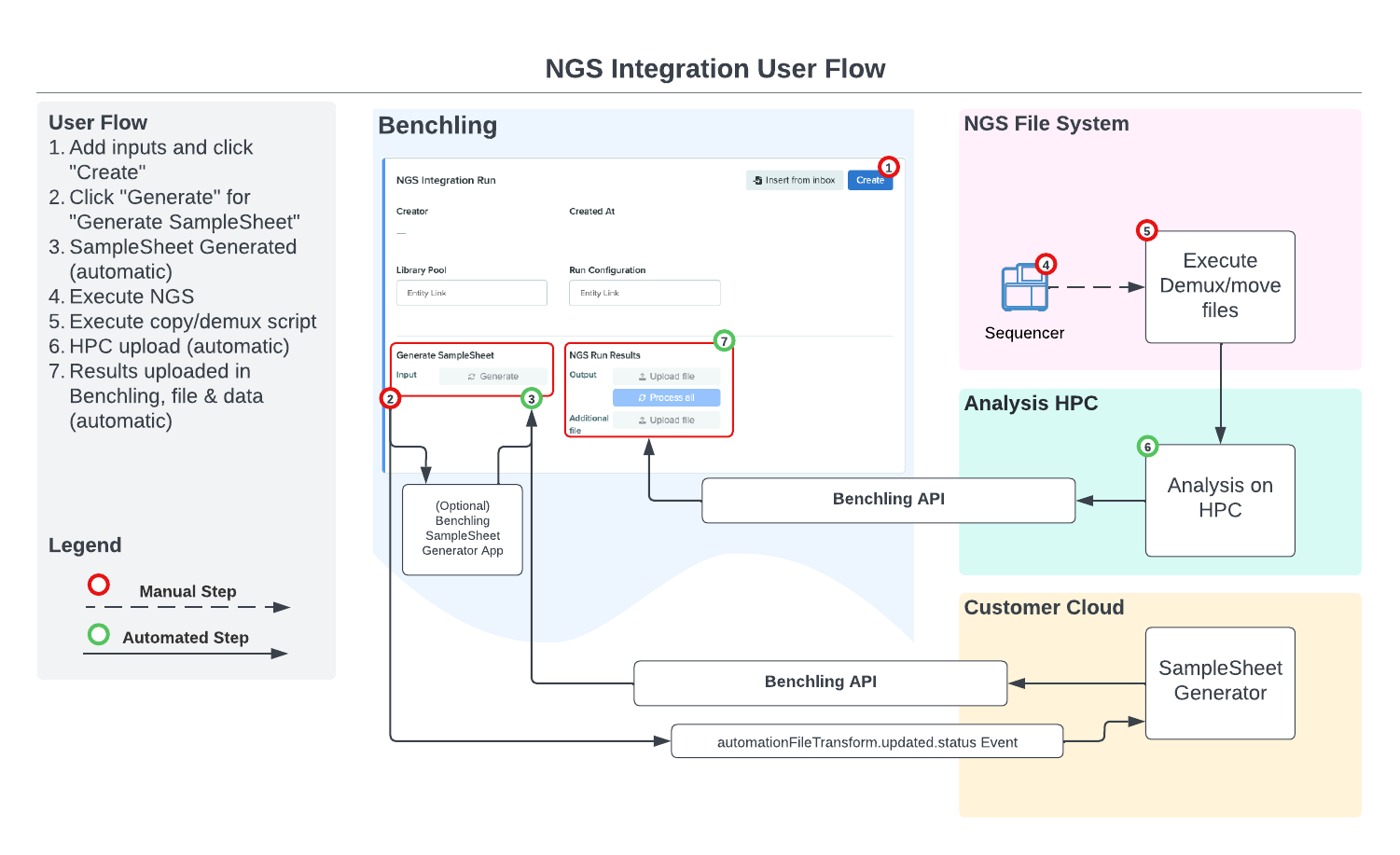
App Canvas
App Canvases provide the smoothest user experience, through an investment of developer time and maintenance. Additionally, Benchling has developed a few apps that can be sold separately to cut out the development time and maintenance costs that come with this approach. For example, a SampleSheet Generation App exists today. In this version of the document, the below sections will detail a technical solution that uses a combination of Benchling Connect and App Canvas. Benchling Connect will be used to process inputs and outputs of file-based
operations related to Benchling's Registry and Inventory, while App Canvases will help the integration smoothly communicate statuses and messages back to the end user. Please refer to our Introduction to App Canvas and Introduction to App Status guides for more information about creating an App Canvas to use with
this integration.
Step by Step
SampleSheet Generation
This integration will be driven through the use of a Benchling Connect run placed inside the user's execution notebook entry. The inputs and outputs will be contained within this object: Run inputs, as described below, will be placed into the run fields, while outputs will appear in the processors at the bottom of the run object.
Inputs
In order for the NGS SampleSheet Generator to execute, the following data and delivery must be executed:
Data
-
A Library Pool entity, containing linkages to indexed libraries and upstream samples.
-
A Run Configuration entity, containing parameters with which the NGS Processing should be run.
- Also consider using run fields for configuration points. If the configurations are less likely to change between runs, use an entity.
-
SampleSheet Data, generated by the automation input generator
- Automation Input Generation will be used to look up necessary data about all entities included in this run, including sample name, index IDs, indexes, and other run-specific field values.
Delivery
-
User clicks "Create" on the Connect Run after completing all necessary run fields
-
User clicks "Generate" on the Input Generator, labeled "SampleSheet Data“
Processing
-
The integration should receive an
[v2-beta.automationInputGenerator.completed](https://docs.benchling.com/docs/events-reference#v2-betaautomationinputgeneratorcompleted)event and accept the event payload -
The integration should pull the Run Configuration entity and the file generated from the Run via API.
-
The Integration should add the
settingssection anddataheader to the file pulled in step 2. This file should then be uploaded to Benchling and the "SampleSheet" Run field should be updated with the file's Blob ID.
Outputs
As a result of this integration, the following responses are expected. The user will click "re-sync" on the Run to see the results:
- Upon success, a completed SampleSheet.csv is available for download from the "SampleSheet" field on the Run. This file should conform to the format shown above, in "SampleSheet Format", and can be used for processing of the NGS data. The user is expected to download this file and provide it in the necessary places in the NGS pipeline.
- An example of a generated SampleSheet can be found below. Please note that different sequencers and pipelines have different requirements for the format of the SampleSheet.
-
Upon error, an error message may be pushed as a Message to the App Canvas. Messages may include:
- "Generated input file not in expected format"
- "Incomplete/invalid run fields"
- "Unexpected error. Please try again."
Recording Analysis Results with Benchling Connect
Upon completion of the NGS pipeline, the results may be recorded against relevant entities back in a Benchling Notebook entry. In this case, the integration will call the Benchling APIs to upload files and results data against schematized results tables. In the UI, the user will view the updated results data or analysis status.
Inputs
For this portion of the integration, the results data will have already been generated and stored within the filestore. This results capture will be triggered by completion of NGS secondary analysis pipelines.
Data
The inputs to this integration are secondary analysis data captured in a per-Sample fashion within the integration.
Data that is written back to Benchling Results may populate one or many Results tables. These schemas can be configured to capture relevant bioinformatics results, like QC status, confirmation results, quality scores, and others depending on the goals of your analysis.
Processing
The delivery of data back to Benchling will target Benchling Results schemas and Assay Run objects.
Results Upload
-
After completion of the analysis pipelines, prepare and upload a tabular file (.csv,.xlsx) with Results data. The results values will be mapped according to Benchling configuration. The results files can be uploaded with the Upload Blobs API endpoint.
-
Create a processor in the Assay Run object that is on the user's notebook entry using the Create Automation Output Processor endpoint. This endpoint requires an
assayRunId, which is a value that the integration will have to persist from the event payload captured during the processing of the SampleSheet Generation portion of the integration. -
Call the Process Output endpoint to automatically process the data from the upload and relate the data to Benchling entities.
Run Object Update
Once the data has been uploaded and processed, you may desire to write a status message to the run object so that the user knows to review the data upload. This can be accomplished with App Status messages
Examples of typical messages may include:
- "NGS Results upload complete. Please review your data."
- "Unexpected error occurred. Please contact Integration support."
Outputs
Upon completion of the results upload, the user may review the status message and results in structured tables in the run object within their notebook entry.
NGS Run Monitoring
As the NGS pipeline progresses from the beginning through primary and secondary analyses, the integration may choose to push updates back to the Benchling UI. This reduces the amount of travel the user has to do to understand the status of their sequencing run, which can span hours.
Processing
Work with the customer development team to define the appropriate triggers for an update to be sent to Benchling. Common triggers/events include:
- Sequencing initiated/in progress
- Sequencing Complete
- QC/demux in progress
- File Transfer in progress
- Secondary Analysis in progress
- NGS Pipeline complete
In order to act on these triggers, the developer may construct a simple script/subroutine to push updates to Benchling, and call that during the execution of a master script, or from within their pipelining tool.
Monitoring status messages should be pushed via App Status messages to the App Canvas that is being used on the associated Run.
Updated 8 months ago
