Use APIs to process files with Lab automation
Overview
This guide demonstrates how to leverage Benchling's API endpoints to:
- Generate Input files
- Insert Output files
- Process (multiple) Lab Auto runs within Notebook Entries
This is a summary of the configurations involved:
- Notebook (NB) Entry that has been pre-populated with Run(s), eg:
RunA,RunB,RunC - Each of the Runs belong to a different run schema (see docs on how to get the IDs):
- RunA schemaId:
assaysch_pV6XCooH - RunB schemaid:
assaysch_rF5GHolP - RunC schemaId:
assaysch_PmvUoRMA
- RunA schemaId:
- Each of the runs contain some
runfieldsandoutputfileconfig(s)which ingest files (eg: results from an instrument) and perform the following actions in Benchling:- Register entities
- Add results against the entities
- Transfer entities to a different location
Workflow Walkthrough
A1. First thing we need is the user creating an Entry and start to work on the runs.
Assumptions:
→ NB entry being created from a template (can even be an request execution entry)
→ All the runs in the NB entry are different “Run Schemas”
Reason: We see this case most frequently so we are going to go ahead with that
- User starts to work on the entry and adds information for the entities (samples, result table, reg tables etc)
- User navigates to run section(s) and clicks on “+” → “create run”. Lets call these RunA, RunB, RunC (in that order in the NB)
- If any fields are present for the run, the user fills them in and then creates the run
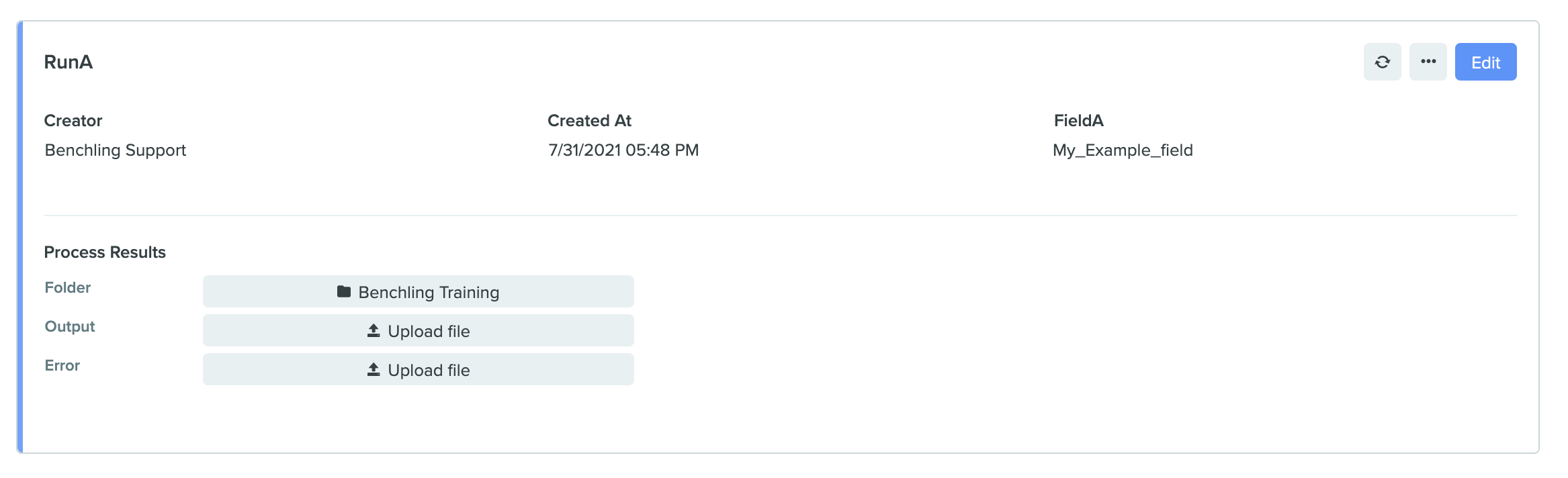
RunA example
- User continues their work in the meanwhile we can look at how API will now interact with this in the steps below
A2. Get the RunId & Entry ID which the user is working on.
Assumptions:
→ Runs have already been created in the NB entry (this step must be done manually in the entry or by creating a run using this endpoint and manually adding it from the inbox).
- One can do that by using the GET command:
yourtenantName.benchling.com/api/reference#/Assay%20Runs/listAssayRunsand providing the SchemaId option (will be something likeassaysch_yzFj9plv) - The return json has a field for
EntryId. Will look something like this:etr_uMs6tSHK - The return json also has a field for the Run
ID. Will look something like this:d9242e5a-9abc-4209-8e87-65eb2f765c00which we can now use for getting information about the run(s) and perform actions (below) - This way one can get the RunId for all the runs (runA, runB, runC) across multiple notebook entries.
B(X). Input File generation
(optional step, slight detour. Skip to Step B1 to continue foroutputfileprocessing )
Let’s say the run you are interested in has an input file generation process. Follow the workflow mentioned below to create that file using the API
Assumptions:
→ Run has already been created
- Go to the Endpoint
yourtenantName.benchling.com/api/reference#/Assay%20Runs/listAutomationInputGeneratorsand provide the assay_run_id (from the step A2 above) - The response json will contain the ID of the automation-input-generator. Something like this:
"id": "aif_cvPqFaXD" - Now navigate to this endpoint:
yourtenantName.benchling.com/api/reference#/Lab%20Automation/generateInputWithAutomationInputGeneratorto generate an input file.- provide the input_generator_id (from the step #2 above)
“aif_cvPqFaXD”
- provide the input_generator_id (from the step #2 above)
- This API call generated a long running task and return json will have a “taskId” for it. Something like this:
“3fa85f64-5717-4562-b3fc-2c963f66afa6" - Now you can navigate to this GET endpoint to check the status of this task:
yourtenantName.benchling.com/api/reference#/Tasks/getTask - There you an provide the taskId and check the status of the task in return json (RUNNING, SUCCEEDED, FAILED, etc)
NOTE
- The “status” field (next to “id”) in the response json has to be “SUCCEEDED”, eg:
{"id": "aif_cvPqFaXD", "status": "SUCCEEDED"}- It is not the “status” at the end of the json (which reflect the overall status of creation of the automation output processor)
- Failures here can arise from incorrect files, errors in the automation runs.
- After the task has SUCCEEDED, the generated input file can be downloaded as a blob which can be found in the response json from task above. Will look something like this:
"file": {
"id": "d3a00259-6cdb-4c4f-94bd-544c94db57fe",
"mimeType": "text/csv",
"name": "file_996d9762-b647-449d-8cd3-14ecf2c29bd2_2021-07-05T15.05.41Z.csv",
"type": "RAW_FILE",
"uploadStatus": "COMPLETE"
}- Navigate to the endpoint:
yourtenantName.benchling.com/api/reference#/Blobs/getBlobUrl - provide the “id” from the response json above (
d3a00259-6cdb-4c4f-94bd-544c94db57fe) - The response json will contain a field “downloadURL” with a value to an amazon s3 bucket that can be visited to download the file. Will look something like this:
{"downloadURL": "https://benchling-results.s3.amazonaws.com/blobs/d3a00259-6cdb-4c4f-94bd-544c94db57fe/Ordering_Primers_996d9762-b647-449d-8cd3-14ecf2c29bd2_2021-07-05T15.05.41Z.csv?response-content-disposition=attachment%3Bfilename%3D%22Ordering%20Primers_996d9762-b647-449d-8cd3-14ecf2c29bd2_2021-07-05T15.05.41Z.csv%22&response-content-type=text%2Fcsv&X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Expires=3600&X-Amz-Date=20210705T152127Z&X-Amz-SignedHeaders=host&X-Amz-Security-Token=IQoJb3JpZ2luX2VjEEUaCXVzLWVhc3QtMSJIMEYCIQCnR6MsPWp2P0olpBMIb4rpSzc2ZBEngoJp27l6Qw34AAIhAOhv2h5PlTqbpR%2BkKOFENscFBODLtUhSQ4hP8f22IZT6Kv8DCC4QABoMMzcxNTg5NjU2MDY4Igy%2BaJFcV794oEEg42Mq3ANIIOftXIyftav16LlySxat4ltmawHm53guF%2BS1AqNXw%2FKbtu6GTIxcYeKrSDtSfbWzQ%2FND305eJ3SMToLPjaCMtDiz2SHHLkEeV49MVxdYo91DeuNpC1dQ7lPmdjrgP9oVZyLFj5kAFQdlevt%2BP9JkxdyEUICkxumKUm9wyIjoGlAZd0xg12MyYpDNPdN%2F6U6iTtjaohIUjcBuRHl28a6xvkqxjK4ILpOYJbluuk0LKIqWO8uRLcLHxacKQuWl5zmPElra4m17%2FWNU6WseOGdtRBUIp%2BxV1dXdu0toe2zjUn1P9dlrqQwJkjZQEzwccCwwIC6h%2F1HVlVm8Vn3gyGFP0kf6AkVlh8%2BMxe%2BN7E15D6yVsKxTuZ88HnUhZxIpt%2BwGVFnDL6S3kCaDP5QsIWGPFkNSpsMnOjM0PqT2Il8R9o42hzxvDpPIqZhsc%2Fbu7GL3hIQaH1S67%2Bw%2B0N1skKEyDBt2Rm6ZHhPV3jZWVSBEXnuj0F7thmO4xyWR4i%2F2yGh5jtm6qfdPM9C2qLA0hy1a8EEnvpHzfixUVa5IOqevOpkKzJxl7VLc7Spg8lZX8bIc%2FU6oWOUPbUs9XCtQmQHL2nEFyPwZWJkV30dgHXL1FJdpGR0HOvgcv6ICZTCa%2FouHBjqkAYvk6BSGGI4kumw69gMgmsG%2BZBb68B%2FGVVfAJ6sjLIEP2m%2B8AwBrhtWfko0VWfrnHnSyNfDLEWGmoFhrIsGKO%2FB%2BCBW9qio1lFL6RbfTkdRwQVodsdpTPl1PwKI3qTEVhu9KTiWDP%2BmgFEKXRFDtOb4vINUtN9oF9lgg%2BpIGAg09PTW1Z5cRMJ40RjU95IVO00XVMXHRnl41y778HENCz8TF28Eq&X-Amz-Credential=ASIAVNBDYFYCI7XPISGC%2F20210705%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Signature=692e182cd267dbb7a3990f3f45167",
"expiresAt": 1625502087}- You can use a file downloader library of your choice for getting the file from this URL (it’s an s3 link)
- The file can be transferred to an instrument / external system for ingestion.
- (optional) Click on the “refresh icon” ♻️ to refresh the run inside the NB and view the generated Input file in the UI.
B1. Prepare the CSV file(s) for upload to Benchling
Now that we have the Entry (id), Run (Id) we can start to send information for files coming from instruments / other systems in those run(s).
Assumptions:
→ The CSV files already are available
→ the matching of which CSV file to use for which Run/Entry combination is already done (eg: using the sample information in result tables, execution of a request etc.)
→ The CSV file is less that 10 MB, if it is greater than 10 MB, the flow is similar, but requires doing a multi-part blob upload
- In order to upload files (blobs) to Benchling, first the blob (file) needs to be converted into base64-encoding. This can be achieved by a simple base64 command in your terminal and providing the csv file as parameter eg:
base64 example.csv - This will return a really long string. Something like this:
IQESyDICfuV5+9bq7LlzskfjqjmxTE+X+fk+qu32vO3DKxD9gyo+pUuVckIMvvkePnxEvftjRpxSip/7pke4ZPeePZJfn/JESCU1w7Wx0gKxefuEY2ppU9uPpxwhgseOHZPCOvFXuFCh1JKa/Zhc3aOraM6fP28mCu3Tj+6TwAkcY2PjlEvJVB/xh5jHxcWaTgLlsJOYCGd99NkQ0yFhPbevIS6PMsTFxUspHQFk5OcGfPPgZxIggcwR8KsJS9+iY2219bJ9j6X2Gd6p9VBTSwNhzYy4Is5cuWLF1LJz9mfk2k7iVDYgmOgg0uok3KfCCy+jHVVahuWN6aXB+e98+JHM1TmHp3RkUV1/BiBKJyaxsgfCDcOa+JQMXrj7MfuU0nAfCZBA1hLwa/HO2qoyt7QIYHSxXh9cwrrx/gOfS5YUsfxHH3wg2X7uIAESuDwEAlq8Q5QZVj1g3XHRwqmHMy4P2sC6KkYXPwwbqj - Next , we also need a md5 for the same file (blob), This can be done by running something like:
md5 example.csvin the terminal - This will return a value such as:
0e03d2cced758fb10faff5833d4a1f06 - Now we can upload this blob to Benchling by navigating to this POST endpoint
yourtenantName.benchling.com/api/reference#/Blobs/createBlob - There one can provide the generated string above as value to parameter for “data64” and providing the value for parameter md5 from the step above as well as providing the:
mimetype (filetype), name (name of the file) & type (RAW_FILE OR VISUALIZATION) - The final post will look something like this:
{
"data64": "IQESyDICfuV5+9bq7LlzskfjqjmxTE+X+fk+qu32vO3DKxD9gyo+pUuVckIMvvkePnxEvftjRpxSip/7pke4ZPeePZJfn/JESCU1w7Wx0gKxefuEY2ppU9uPpxwhgseOHZPCOvFXuFCh1JKa/Zhc3aOraM6fP28mCu3Tj+6TwAkcY2PjlEvJVB/xh5jHxcWaTgLlsJOYCGd99NkQ0yFhPbevIS6PMsTFxUspHQFk5OcGfPPgZxIggcwR8KsJS9+iY2219bJ9j6X2Gd6p9VBTSwNhzYy4Is5cuWLF1LJz9mfk2k7iVDYgmOgg0uok3KfCCy+jHVVahuWN6aXB+e98+JHM1TmHp3RkUV1/BiBKJyaxsgfCDcOa+JQMXrj7MfuU0nAfCZBA1hLwa/HO2qoyt7QIYHSxXh9cwrrx/gOfS5YUsfxHH3wg2X7uIAESuDwEAlq8Q5QZVj1g3XHRwqmHMy4P2sC6KkYXPwwbqj",
"md5": "0e03d2cced758fb10faff5833d4a1f06",
"mimeType": "text/csv",
"name": "my_instrument_csv_file_runA.csv",
"type": "RAW_FILE"
}- In the response json, one can get the blob id with the field: ID which will look something like this:
c33fe52d-fe6a-4c98-adcd-211bdf6778f7 - This is the ID which we will need to process the Lab automation run in the next steps.
Don't lose your blob IDIf you lose track of your blob ID, it cannot be looked up in Benchling and the file will need to be re-uploaded. So make sure to save it!
This step (B1) can be repeated for all files for each of the run(s)
C. Process the run(s) with the uploaded CSV file(s)
Now that we know the Run Id(s), have the Blob ID(s), we can start to process each of the run using the respective blob(s).
Assumptions:
→ The CSV files already have been uploaded to Benchling and we have the Blob IDs (from step B1 above)
C1. Create Automation Output Processor
- Navigate to this POST endpoint:
yourtenantName.benchling.com/api/reference#/Lab%20Automation/createAutomationOutputProcessorand provide the respective parameters:assayRunID: ID of the assay run from step BautomationFileConfigName: Name of the output processor (UI name)fileId: this is the blob id from Step C
- The call would look something like this:
{
"assayRunId": "3da3a41b-3e92-4551-9c8b-644d5f465268"
"automationFileConfigName": "1A. Process file",
"fileId": "c33fe52d-fe6a-4c98-adcd-211bdf6778f7"
}- In the response json, once can see the ID for this particular outputProcessor, this will look something like this:
"id": "aop_C3wGA9HF"
C2. Process the generated Automation Output Processor
Now that the automation Output Processor has been created, we can go ahead and “Process” it.
- Navigate to this POST endpoint:
yourtenantName.benchling.com/api/reference#/Lab%20Automation/processOutputWithAutomationOutputProcessorand provide the respective parameters:
* output_processor_id : id of created automation output process in step D1 above
(eg:"aop_C3wGA9HF") - this API call generated a long running task and return json will have a “taskId” for it. Something like this:
“3fa85f64-5717-4562-b3fc-2c963f66afa6" - Now you can navigate to this GET endpoint to check the status of this task:
YourtenantName.benchling.com/api/reference#/Tasks/getTask - There you an provide the taskId and check the status of the task in return json (RUNNING, SUCCEEDED, FAILED etc.)
NOTE
- In case of errors one can archive the created
automationoutputprocessorusing this endpoint:yourtenantName.benchling.com/api/reference#/Lab%20Automation/archiveAutomationOutputProcessors- Followed by which, one can redo the previous step by using this endpoint:
yourtenantName.benchling.com/api/reference#/Lab%20Automation/updateAutomationOutputProcessorby Providing theoutput_processor_idand thefileId(example if you have uploaded a new file (blob))
The steps C1 & C2 can be repeated for all of the respective Run(s)
D. View the processed run(s) in NB entry
Assumptions:
→ The task from Step C2 shows “status” (for automation output processing) as “SUCCEEDED"
- After all the steps above (A-C) have been completed, one can navigate to the original NB entry and view / inspect the run
- click on the “refresh icon” ♻️ to refresh the run and view the generated results from output file processing.
Steps A-D can be repeated for all runs, across multiple NB entries
If you are looking to monitor the success of this process or keep track of which runs have been processed in which NB entries, you can log aresult with that information and track it in Benchling.
Updated about 1 year ago
